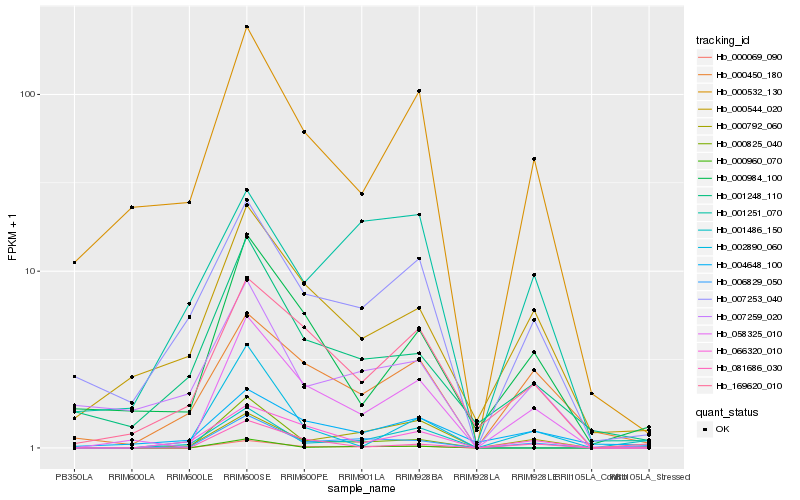
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006829_050 |
0.0 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 2 |
Hb_058325_010 |
0.2258256179 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 3 |
Hb_001248_110 |
0.2369538646 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 4 |
Hb_000960_070 |
0.2497488939 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 5 |
Hb_000792_060 |
0.2501053692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000544_020 |
0.2518435411 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 7 |
Hb_004648_100 |
0.2607394605 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 8 |
Hb_007253_040 |
0.2613794353 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 9 |
Hb_169620_010 |
0.2615866384 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 10 |
Hb_066320_010 |
0.2654612775 |
- |
- |
hypothetical protein VITISV_027639 [Vitis vinifera] |
| 11 |
Hb_000450_180 |
0.2670306794 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 12 |
Hb_001486_150 |
0.2672306939 |
- |
- |
- |
| 13 |
Hb_000984_100 |
0.2674350721 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 14 |
Hb_007259_020 |
0.2693103348 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 15 |
Hb_000069_090 |
0.2746844649 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 16 |
Hb_081686_030 |
0.2764471675 |
- |
- |
hypothetical protein MIMGU_mgv11b016770mg [Erythranthe guttata] |
| 17 |
Hb_000825_040 |
0.2808276819 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_002890_060 |
0.2837213895 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 19 |
Hb_001251_070 |
0.2849392255 |
- |
- |
PREDICTED: probable aldo-keto reductase 2, partial [Populus euphratica] |
| 20 |
Hb_000532_130 |
0.2867242217 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |