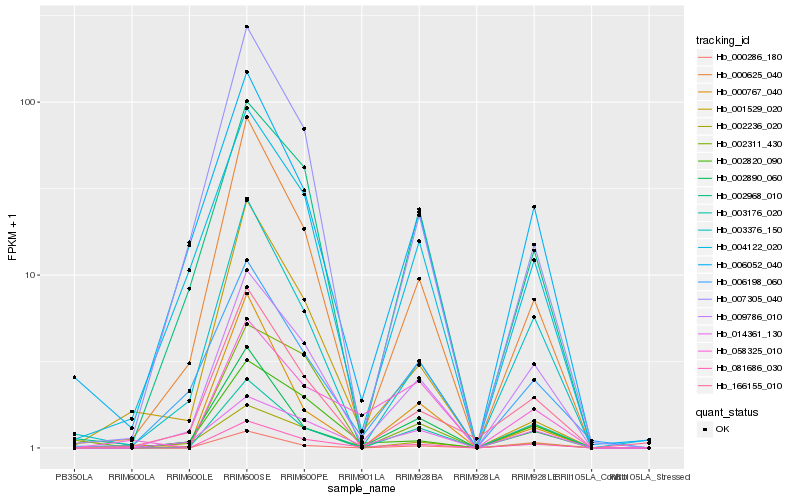
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_081686_030 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv11b016770mg [Erythranthe guttata] |
| 2 |
Hb_009786_010 |
0.1275680558 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 3 |
Hb_000625_040 |
0.1399893634 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 4 |
Hb_002820_090 |
0.1602142514 |
- |
- |
PREDICTED: WAT1-related protein At1g21890-like [Jatropha curcas] |
| 5 |
Hb_166155_010 |
0.1707063854 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 6 |
Hb_003176_020 |
0.1717728373 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 7 |
Hb_003376_150 |
0.1744338683 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 8 |
Hb_058325_010 |
0.1831902088 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 9 |
Hb_007305_040 |
0.186353961 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 10 |
Hb_002890_060 |
0.1893167793 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 11 |
Hb_002968_010 |
0.1940734845 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 12 |
Hb_002311_430 |
0.1960348632 |
- |
- |
hypothetical protein JCGZ_18918 [Jatropha curcas] |
| 13 |
Hb_000286_180 |
0.1966171825 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |
| 14 |
Hb_001529_020 |
0.1986491329 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_014361_130 |
0.1988738729 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 16 |
Hb_002236_020 |
0.2003060903 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 17 |
Hb_000767_040 |
0.2009817586 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_004122_020 |
0.2031395432 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 19 |
Hb_006198_060 |
0.2051321036 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 20 |
Hb_006052_040 |
0.2056364406 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |