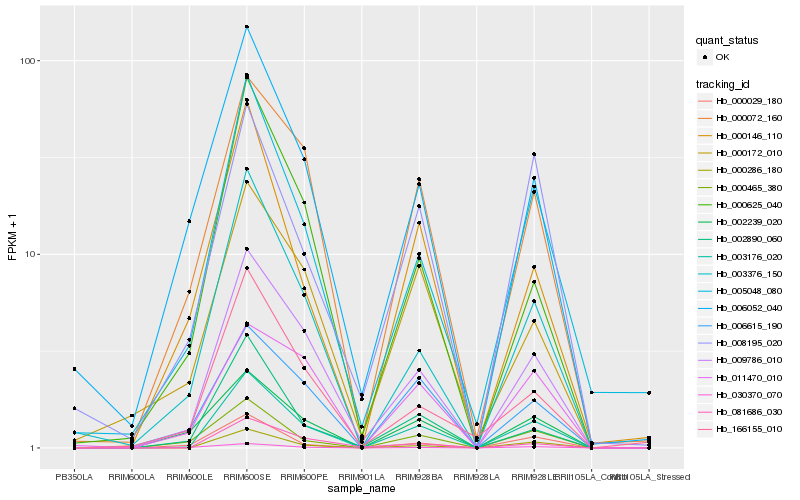
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003176_020 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 2 |
Hb_000286_180 |
0.1099913589 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |
| 3 |
Hb_009786_010 |
0.1136930321 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 4 |
Hb_003376_150 |
0.1345177901 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 5 |
Hb_005048_080 |
0.1429217413 |
- |
- |
PREDICTED: PAP-specific phosphatase HAL2-like [Jatropha curcas] |
| 6 |
Hb_000072_160 |
0.1575148882 |
- |
- |
proline oxidase [Manihot esculenta] |
| 7 |
Hb_000465_380 |
0.1638077391 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_006615_190 |
0.1682044602 |
- |
- |
UDP-glycosyltransferase 1 [Linum usitatissimum] |
| 9 |
Hb_002890_060 |
0.169467765 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 10 |
Hb_006052_040 |
0.1711601531 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 11 |
Hb_081686_030 |
0.1717728373 |
- |
- |
hypothetical protein MIMGU_mgv11b016770mg [Erythranthe guttata] |
| 12 |
Hb_008195_020 |
0.1720066763 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 13 |
Hb_000029_180 |
0.1725558096 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 14 |
Hb_000625_040 |
0.1750520773 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 15 |
Hb_002239_020 |
0.1755411746 |
- |
- |
MLO, putative [Theobroma cacao] |
| 16 |
Hb_000172_010 |
0.1767336237 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000146_110 |
0.1779662899 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 18 |
Hb_011470_010 |
0.1828742428 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 19 |
Hb_166155_010 |
0.1831520645 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 20 |
Hb_030370_070 |
0.1838953703 |
- |
- |
ATP binding protein, putative [Ricinus communis] |