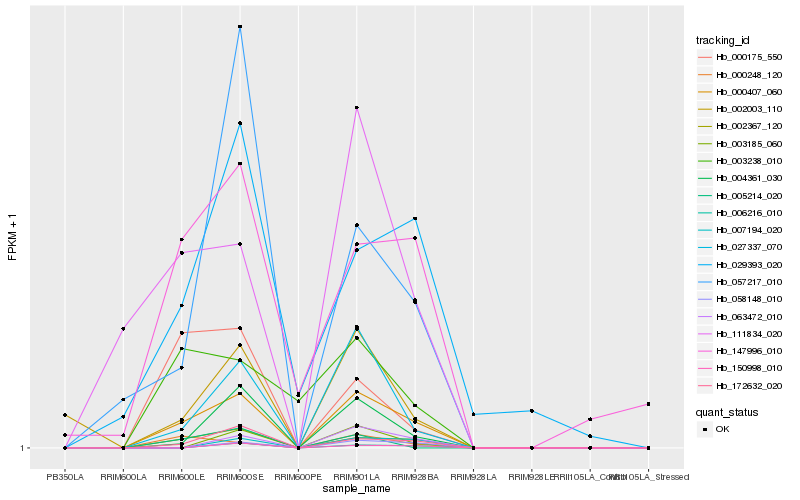
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000407_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_002367_120 |
0.1265151956 |
- |
- |
- |
| 3 |
Hb_172632_020 |
0.1852662681 |
- |
- |
- |
| 4 |
Hb_027337_070 |
0.1944412464 |
- |
- |
hypothetical protein POPTR_0001s29030g [Populus trichocarpa] |
| 5 |
Hb_004361_030 |
0.2260883831 |
- |
- |
PREDICTED: uncharacterized protein LOC104239397 isoform X1 [Nicotiana sylvestris] |
| 6 |
Hb_000248_120 |
0.2420053139 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 7 |
Hb_057217_010 |
0.2548187069 |
- |
- |
PREDICTED: uncharacterized protein LOC104594412 [Nelumbo nucifera] |
| 8 |
Hb_002003_110 |
0.2550215311 |
- |
- |
PREDICTED: uncharacterized protein LOC105634406 [Jatropha curcas] |
| 9 |
Hb_111834_020 |
0.2595553722 |
- |
- |
PREDICTED: protein phosphatase methylesterase 1 [Jatropha curcas] |
| 10 |
Hb_000175_550 |
0.2615966577 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 11 |
Hb_147996_010 |
0.2792756451 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003238_010 |
0.2854165754 |
- |
- |
PREDICTED: uncharacterized protein LOC104880636 [Vitis vinifera] |
| 13 |
Hb_029393_020 |
0.2928276526 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 14 |
Hb_005214_020 |
0.2990398794 |
- |
- |
hypothetical protein JCGZ_12886 [Jatropha curcas] |
| 15 |
Hb_058148_010 |
0.2999216357 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 16 |
Hb_063472_010 |
0.300296237 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |
| 17 |
Hb_003185_060 |
0.3035471588 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Amborella trichopoda] |
| 18 |
Hb_007194_020 |
0.3044731864 |
- |
- |
PREDICTED: uncharacterized protein LOC105641615 [Jatropha curcas] |
| 19 |
Hb_006216_010 |
0.3045394182 |
- |
- |
PREDICTED: uncharacterized protein LOC103411650 [Malus domestica] |
| 20 |
Hb_150998_010 |
0.3102743397 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |