| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
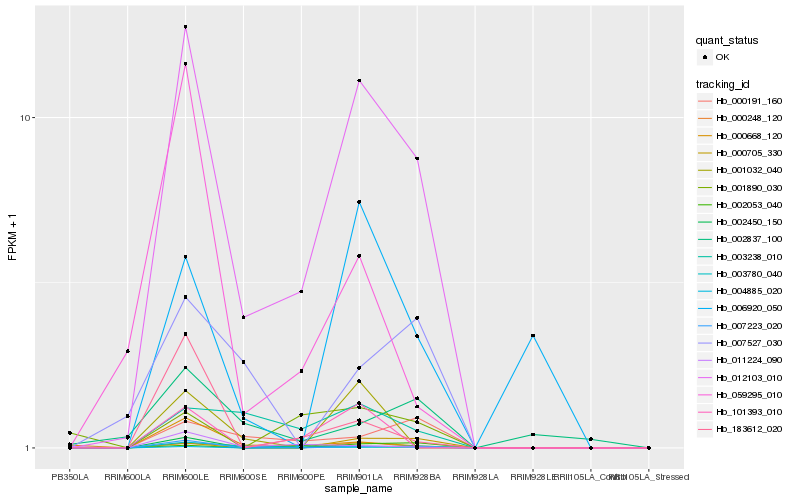
Hb_012103_010 |
0.0 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 2 |
Hb_000248_120 |
0.2571998673 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 3 |
Hb_000191_160 |
0.2927868889 |
- |
- |
hypothetical protein EUGRSUZ_C03547 [Eucalyptus grandis] |
| 4 |
Hb_007223_020 |
0.2940500347 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-3 [Jatropha curcas] |
| 5 |
Hb_003238_010 |
0.2962295142 |
- |
- |
PREDICTED: uncharacterized protein LOC104880636 [Vitis vinifera] |
| 6 |
Hb_004885_020 |
0.3098836747 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_003780_040 |
0.3194719364 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 8 |
Hb_000668_120 |
0.3213247034 |
- |
- |
hypothetical protein JCGZ_11991 [Jatropha curcas] |
| 9 |
Hb_007527_030 |
0.3300972424 |
- |
- |
- |
| 10 |
Hb_002450_150 |
0.3364720022 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_183612_020 |
0.3373649906 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 12 |
Hb_006920_050 |
0.3397518587 |
- |
- |
endo-1,4-beta-glucanase [Vigna radiata var. radiata] |
| 13 |
Hb_059295_010 |
0.340683106 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 14 |
Hb_002053_040 |
0.3443670039 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 24 [Jatropha curcas] |
| 15 |
Hb_000705_330 |
0.3470604532 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 16 |
Hb_101393_010 |
0.3473372798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002837_100 |
0.3517547223 |
- |
- |
PREDICTED: F-box protein PP2-B15-like [Jatropha curcas] |
| 18 |
Hb_001032_040 |
0.3606190842 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001890_030 |
0.3626659633 |
- |
- |
- |
| 20 |
Hb_011224_090 |
0.3635312523 |
- |
- |
PREDICTED: ABC transporter G family member 26 [Jatropha curcas] |