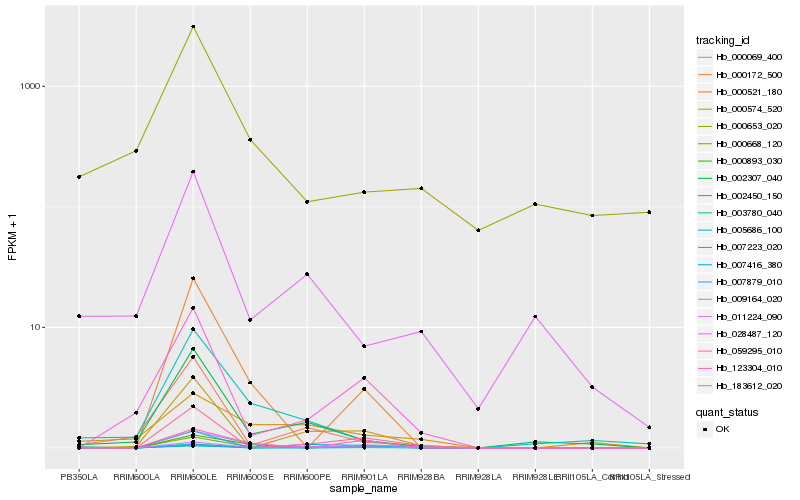
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000668_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_11991 [Jatropha curcas] |
| 2 |
Hb_011224_090 |
0.1402945134 |
- |
- |
PREDICTED: ABC transporter G family member 26 [Jatropha curcas] |
| 3 |
Hb_007879_010 |
0.2276316936 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 4 |
Hb_003780_040 |
0.2426569511 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 5 |
Hb_183612_020 |
0.2482911492 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 6 |
Hb_009164_020 |
0.2635401673 |
- |
- |
hypothetical protein PRUPE_ppa026212mg, partial [Prunus persica] |
| 7 |
Hb_059295_010 |
0.2649323244 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 8 |
Hb_002450_150 |
0.2771457102 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000574_520 |
0.2790720213 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 10 |
Hb_028487_120 |
0.2903974583 |
- |
- |
PREDICTED: HMG-Y-related protein B-like [Jatropha curcas] |
| 11 |
Hb_000172_500 |
0.2954575289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002307_040 |
0.2959352117 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000521_180 |
0.3004190393 |
- |
- |
PREDICTED: uncharacterized protein LOC105650335 [Jatropha curcas] |
| 14 |
Hb_000069_400 |
0.3091211929 |
- |
- |
RING-H2 finger protein ATL3K, putative [Ricinus communis] |
| 15 |
Hb_000893_030 |
0.3113430305 |
- |
- |
PREDICTED: uncharacterized protein LOC105851424 [Cicer arietinum] |
| 16 |
Hb_000653_020 |
0.3114012366 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 17 |
Hb_007223_020 |
0.3116715428 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-3 [Jatropha curcas] |
| 18 |
Hb_005686_100 |
0.3118863467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_123304_010 |
0.3130397678 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 20 |
Hb_007416_380 |
0.3132712004 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |