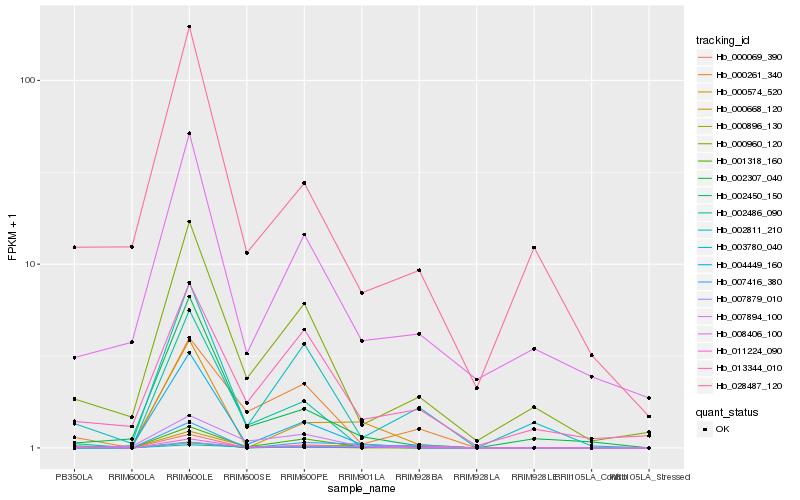
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011224_090 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 26 [Jatropha curcas] |
| 2 |
Hb_000668_120 |
0.1402945134 |
- |
- |
hypothetical protein JCGZ_11991 [Jatropha curcas] |
| 3 |
Hb_001318_160 |
0.2119144237 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 4 |
Hb_000261_340 |
0.2204291835 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 5 |
Hb_002450_150 |
0.2266566092 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_007879_010 |
0.2329113846 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 7 |
Hb_000960_120 |
0.2362798482 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 8 |
Hb_002811_210 |
0.2400703618 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000574_520 |
0.247944094 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 10 |
Hb_013344_010 |
0.2509722657 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 11 |
Hb_002486_090 |
0.2523089547 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |
| 12 |
Hb_003780_040 |
0.2543868 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 13 |
Hb_028487_120 |
0.2558762911 |
- |
- |
PREDICTED: HMG-Y-related protein B-like [Jatropha curcas] |
| 14 |
Hb_000896_130 |
0.2567605043 |
transcription factor |
TF Family: CPP |
cysteine-rich polycomb-like family protein [Populus trichocarpa] |
| 15 |
Hb_007894_100 |
0.2570223085 |
- |
- |
PREDICTED: uncharacterized protein LOC105632952 [Jatropha curcas] |
| 16 |
Hb_002307_040 |
0.2572385722 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_008406_100 |
0.2589215675 |
- |
- |
PREDICTED: protein TPX2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_007416_380 |
0.2593349858 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |
| 19 |
Hb_004449_160 |
0.2598182079 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000069_390 |
0.2599551455 |
- |
- |
PREDICTED: probable F-box protein At3g61730 [Jatropha curcas] |