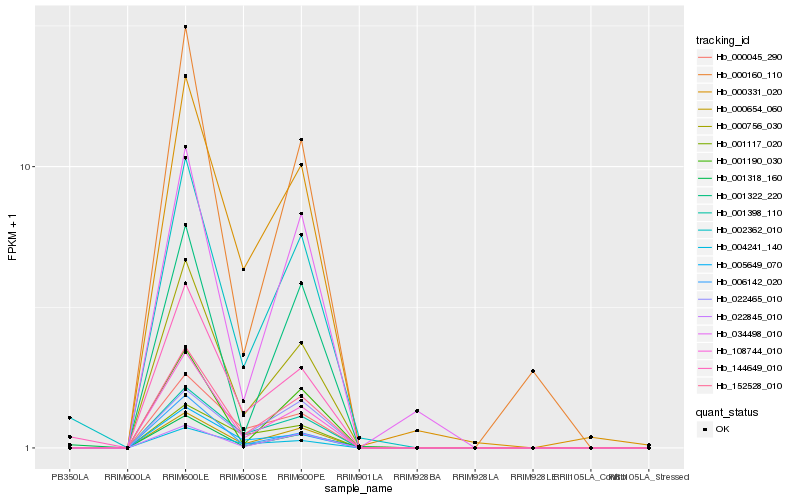
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002362_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_12328 [Jatropha curcas] |
| 2 |
Hb_144649_010 |
0.0959831108 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |
| 3 |
Hb_000756_030 |
0.1097680285 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000654_060 |
0.1098951206 |
- |
- |
hypothetical protein CICLE_v10027550mg [Citrus clementina] |
| 5 |
Hb_152528_010 |
0.1126570526 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 6 |
Hb_108744_010 |
0.1285509103 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_022845_010 |
0.1313254801 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 8 |
Hb_001318_160 |
0.1334032252 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 9 |
Hb_001398_110 |
0.1355207156 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 10 |
Hb_000331_020 |
0.1409509288 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_022465_010 |
0.1487538356 |
- |
- |
hypothetical protein JCGZ_06012 [Jatropha curcas] |
| 12 |
Hb_000045_290 |
0.1490831671 |
- |
- |
- |
| 13 |
Hb_004241_140 |
0.1519401587 |
- |
- |
S-locus lectin protein kinase family protein, putative [Theobroma cacao] |
| 14 |
Hb_001190_030 |
0.1519487564 |
- |
- |
hypothetical protein VITISV_017115 [Vitis vinifera] |
| 15 |
Hb_001322_220 |
0.1562767636 |
- |
- |
Major pollen allergen Ory s 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_005649_070 |
0.1620452605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_034498_010 |
0.1643573156 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 18 |
Hb_001117_020 |
0.1671349051 |
- |
- |
hypothetical protein CICLE_v10008579mg [Citrus clementina] |
| 19 |
Hb_006142_020 |
0.1693116515 |
- |
- |
JHL25P11.12 [Jatropha curcas] |
| 20 |
Hb_000160_110 |
0.1698016654 |
- |
- |
PREDICTED: uncharacterized protein LOC100253584 [Vitis vinifera] |