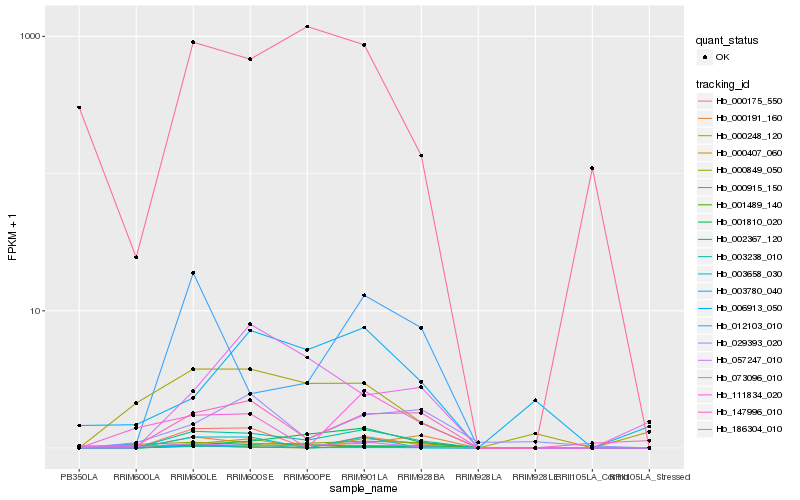
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003238_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104880636 [Vitis vinifera] |
| 2 |
Hb_000248_120 |
0.2685589427 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 3 |
Hb_147996_010 |
0.2742504333 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000407_060 |
0.2854165754 |
- |
- |
- |
| 5 |
Hb_000915_150 |
0.2950268682 |
- |
- |
- |
| 6 |
Hb_012103_010 |
0.2962295142 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 7 |
Hb_000175_550 |
0.3003329726 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 8 |
Hb_057247_010 |
0.3005517381 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 9 |
Hb_002367_120 |
0.3061190427 |
- |
- |
- |
| 10 |
Hb_000191_160 |
0.3089582298 |
- |
- |
hypothetical protein EUGRSUZ_C03547 [Eucalyptus grandis] |
| 11 |
Hb_000849_050 |
0.3134895827 |
- |
- |
hypothetical protein VITISV_010852 [Vitis vinifera] |
| 12 |
Hb_003780_040 |
0.3135511665 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 13 |
Hb_001810_020 |
0.3140656462 |
- |
- |
- |
| 14 |
Hb_186304_010 |
0.3213047118 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 15 |
Hb_029393_020 |
0.3332862225 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 16 |
Hb_006913_050 |
0.339745353 |
- |
- |
- |
| 17 |
Hb_111834_020 |
0.3454221962 |
- |
- |
PREDICTED: protein phosphatase methylesterase 1 [Jatropha curcas] |
| 18 |
Hb_003658_030 |
0.3459413795 |
- |
- |
- |
| 19 |
Hb_073096_010 |
0.3523628293 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 45-like [Jatropha curcas] |
| 20 |
Hb_001489_140 |
0.3579026758 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_21694 [Jatropha curcas] |