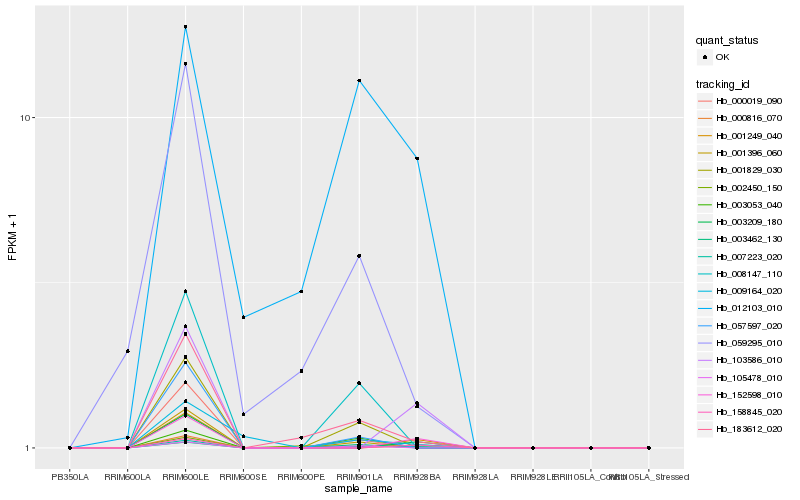
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007223_020 |
0.0 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-3 [Jatropha curcas] |
| 2 |
Hb_183612_020 |
0.2148733789 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 3 |
Hb_001829_030 |
0.2459843138 |
- |
- |
PREDICTED: uncharacterized protein LOC105119087 [Populus euphratica] |
| 4 |
Hb_008147_110 |
0.249773453 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 5 |
Hb_105478_010 |
0.2541915973 |
- |
- |
PREDICTED: uncharacterized protein LOC105762101 [Gossypium raimondii] |
| 6 |
Hb_001396_060 |
0.2562332383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000019_090 |
0.2686780629 |
- |
- |
PREDICTED: thioredoxin H4-1-like [Jatropha curcas] |
| 8 |
Hb_057597_020 |
0.2706014362 |
- |
- |
PREDICTED: anti-H(O) lectin-like [Gossypium raimondii] |
| 9 |
Hb_002450_150 |
0.281075865 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_059295_010 |
0.2844649416 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 11 |
Hb_003209_180 |
0.2918474654 |
- |
- |
Alpha-1,4 glucan phosphorylase L-2 isozyme [Morus notabilis] |
| 12 |
Hb_003053_040 |
0.2925754811 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012103_010 |
0.2940500347 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 14 |
Hb_001249_040 |
0.2950275048 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 15 |
Hb_000816_070 |
0.2980062631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003462_130 |
0.3017478833 |
- |
- |
- |
| 17 |
Hb_152598_010 |
0.302722804 |
- |
- |
PREDICTED: uncharacterized protein LOC104115096 [Nicotiana tomentosiformis] |
| 18 |
Hb_009164_020 |
0.3033400893 |
- |
- |
hypothetical protein PRUPE_ppa026212mg, partial [Prunus persica] |
| 19 |
Hb_103586_010 |
0.3033503367 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Glycine max] |
| 20 |
Hb_158845_020 |
0.3086353418 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |