| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
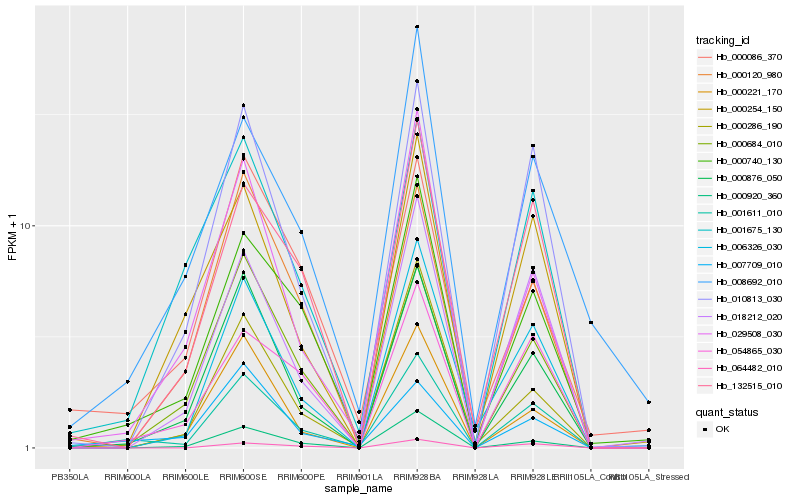
Hb_001611_010 |
0.0 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 2 |
Hb_000740_130 |
0.127147728 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 3 |
Hb_000876_050 |
0.1283312217 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 4 |
Hb_010813_030 |
0.1383321915 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 5 |
Hb_006326_030 |
0.1486690704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_054865_030 |
0.1503363916 |
- |
- |
- |
| 7 |
Hb_018212_020 |
0.1507821128 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 8 |
Hb_132515_010 |
0.1564136467 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 9 |
Hb_029508_030 |
0.1577957478 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0002s04880g [Populus trichocarpa] |
| 10 |
Hb_000684_010 |
0.1582573243 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_007709_010 |
0.1628751955 |
- |
- |
- |
| 12 |
Hb_000920_360 |
0.164605459 |
- |
- |
PREDICTED: (R,S)-reticuline 7-O-methyltransferase-like [Jatropha curcas] |
| 13 |
Hb_064482_010 |
0.1665402683 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_001675_130 |
0.166623947 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 15 |
Hb_000221_170 |
0.1738255809 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 2 [Jatropha curcas] |
| 16 |
Hb_000254_150 |
0.1745196208 |
- |
- |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 17 |
Hb_000120_980 |
0.1755796372 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 18 |
Hb_000086_370 |
0.1783623821 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 19 |
Hb_008692_010 |
0.179550858 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 20 |
Hb_000286_190 |
0.1798969857 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |