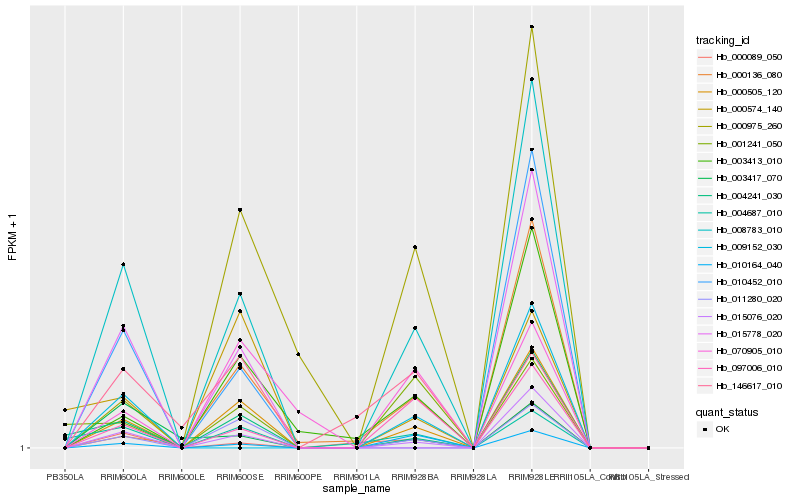
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008783_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000505_120 |
0.1042571253 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_015076_020 |
0.1546313297 |
- |
- |
IMP dehydrogenase/GMP reductase [Hevea brasiliensis] |
| 4 |
Hb_011280_020 |
0.1591431908 |
- |
- |
polyprotein [Oryza australiensis] |
| 5 |
Hb_097006_010 |
0.1802899293 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 6 |
Hb_010164_040 |
0.2231938616 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 7 |
Hb_004241_030 |
0.2347795089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003413_010 |
0.242870036 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 9 |
Hb_000089_050 |
0.2443742124 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_004687_010 |
0.2665450948 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 11 |
Hb_015778_020 |
0.2688892497 |
- |
- |
Putative methyltransferase family protein [Solanum demissum] |
| 12 |
Hb_003417_070 |
0.2723106484 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 13 |
Hb_010452_010 |
0.2769157187 |
- |
- |
PREDICTED: uncharacterized protein LOC104232369 [Nicotiana sylvestris] |
| 14 |
Hb_000136_080 |
0.2896964893 |
- |
- |
hypothetical protein AALP_AA8G499800 [Arabis alpina] |
| 15 |
Hb_070905_010 |
0.2929843436 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 16 |
Hb_001241_050 |
0.2948778359 |
- |
- |
PREDICTED: uncharacterized protein LOC104880586 isoform X1 [Vitis vinifera] |
| 17 |
Hb_000574_140 |
0.2986691953 |
- |
- |
- |
| 18 |
Hb_000975_260 |
0.3065387864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_146617_010 |
0.3109996957 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 20 |
Hb_009152_030 |
0.3131601573 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |