| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
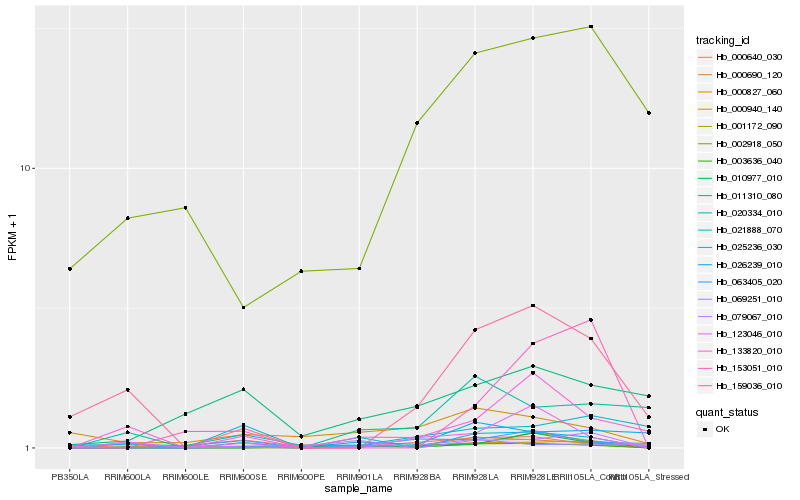
Hb_000940_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 2 |
Hb_003636_040 |
0.1451534622 |
- |
- |
mitochondrial translational initiation factor, putative [Ricinus communis] |
| 3 |
Hb_001172_090 |
0.3418646583 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 4 |
Hb_063405_020 |
0.3455827008 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 5 |
Hb_069251_010 |
0.3530626372 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_000690_120 |
0.3639170373 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_123046_010 |
0.3687480044 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 8 |
Hb_153051_010 |
0.3695152497 |
- |
- |
- |
| 9 |
Hb_159036_010 |
0.3700001588 |
- |
- |
- |
| 10 |
Hb_133820_010 |
0.3759745586 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 11 |
Hb_020334_010 |
0.3770919146 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 12 |
Hb_025236_030 |
0.380201955 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 13 |
Hb_079067_010 |
0.380707445 |
- |
- |
PREDICTED: protein SPT2 homolog [Malus domestica] |
| 14 |
Hb_000640_030 |
0.3808203412 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_021888_070 |
0.3810470909 |
- |
- |
Putative CCR4-associated factor 1 like 11 [Glycine soja] |
| 16 |
Hb_002918_050 |
0.3838859299 |
transcription factor |
TF Family: SBP |
PREDICTED: mitochondrial arginine transporter BAC1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_026239_010 |
0.3840137719 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 18 |
Hb_000827_060 |
0.3845454638 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 19 |
Hb_011310_080 |
0.3852107067 |
- |
- |
hypothetical protein POPTR_0006s26840g [Populus trichocarpa] |
| 20 |
Hb_010977_010 |
0.3875223198 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |