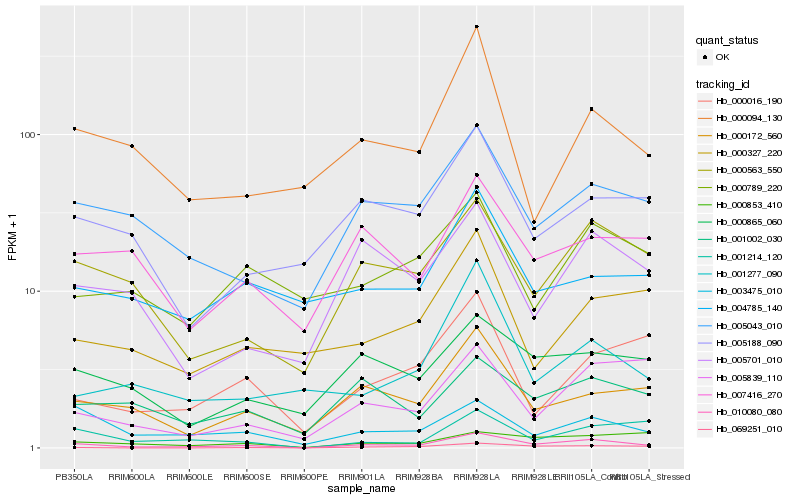
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010080_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000563_550 |
0.1909157182 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 3 |
Hb_005043_010 |
0.2006916173 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_000172_560 |
0.202831304 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Jatropha curcas] |
| 5 |
Hb_003475_010 |
0.2093487718 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 6 |
Hb_000865_060 |
0.2149858741 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |
| 7 |
Hb_005701_010 |
0.2187107187 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001002_030 |
0.2222042356 |
- |
- |
PREDICTED: uncharacterized protein LOC105643537 [Jatropha curcas] |
| 9 |
Hb_000094_130 |
0.2233886093 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 10 |
Hb_005188_090 |
0.2244870743 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 11 |
Hb_007416_270 |
0.2263038037 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 12 |
Hb_001214_120 |
0.2265683707 |
- |
- |
- |
| 13 |
Hb_001277_090 |
0.231145929 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 14 |
Hb_005839_110 |
0.2328839478 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Populus euphratica] |
| 15 |
Hb_069251_010 |
0.2336974207 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_000016_190 |
0.2365918009 |
- |
- |
ABC transporter B family member 28 [Glycine soja] |
| 17 |
Hb_000327_220 |
0.2383733816 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 18 |
Hb_000853_410 |
0.2387889422 |
- |
- |
- |
| 19 |
Hb_004785_140 |
0.2388436993 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000789_220 |
0.2390187474 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |