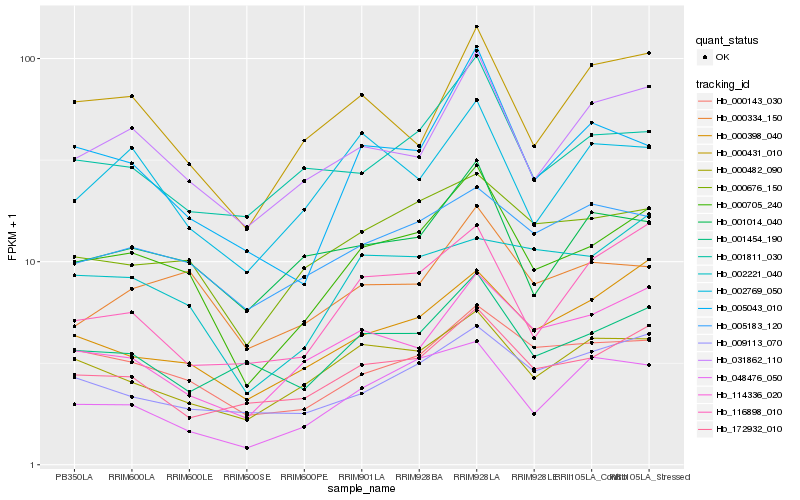
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000705_240 |
0.0 |
- |
- |
DNA-directed RNA polymerase II 19 kD polypeptide rpb7, putative [Ricinus communis] |
| 2 |
Hb_031862_110 |
0.1079322259 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 3 |
Hb_000676_150 |
0.1083442408 |
- |
- |
50S ribosomal protein L7/L12, putative [Ricinus communis] |
| 4 |
Hb_000143_030 |
0.1097547269 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 5 |
Hb_172932_010 |
0.1195529408 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000482_090 |
0.1195578829 |
- |
- |
PREDICTED: uncharacterized protein LOC105638145 [Jatropha curcas] |
| 7 |
Hb_000334_150 |
0.1217957782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000398_040 |
0.1222367127 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 9 |
Hb_005043_010 |
0.1236955833 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_001014_040 |
0.1245244927 |
- |
- |
PREDICTED: uncharacterized protein LOC105647308 [Jatropha curcas] |
| 11 |
Hb_114336_020 |
0.1249339354 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Fragaria vesca subsp. vesca] |
| 12 |
Hb_001811_030 |
0.1268446475 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 13 |
Hb_000431_010 |
0.1282396391 |
- |
- |
hypothetical protein POPTR_0016s05340g [Populus trichocarpa] |
| 14 |
Hb_048476_050 |
0.1309479309 |
- |
- |
semialdehyde dehydrogenase family protein [Populus trichocarpa] |
| 15 |
Hb_005183_120 |
0.1314404502 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 16 |
Hb_001454_190 |
0.1318812263 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g06920 [Jatropha curcas] |
| 17 |
Hb_009113_070 |
0.1324713489 |
- |
- |
hypothetical protein POPTR_0010s07510g [Populus trichocarpa] |
| 18 |
Hb_002769_050 |
0.1335192908 |
- |
- |
PREDICTED: endophilin-A1 [Jatropha curcas] |
| 19 |
Hb_002221_040 |
0.1359090372 |
- |
- |
PREDICTED: uncharacterized protein LOC105639851 [Jatropha curcas] |
| 20 |
Hb_116898_010 |
0.1366931232 |
- |
- |
PREDICTED: uncharacterized protein LOC105639569 isoform X2 [Jatropha curcas] |