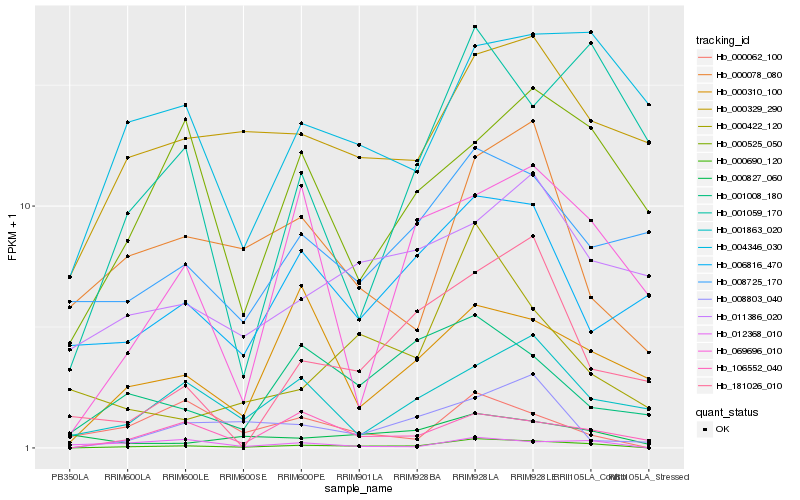
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000690_120 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_106552_040 |
0.2557255004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000329_290 |
0.2757631296 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 4 |
Hb_000078_080 |
0.2798353903 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein JCGZ_17040 [Jatropha curcas] |
| 5 |
Hb_008803_040 |
0.280053927 |
- |
- |
PREDICTED: uncharacterized protein LOC105157106 [Sesamum indicum] |
| 6 |
Hb_001863_020 |
0.2835631017 |
- |
- |
PREDICTED: protein FAM91A1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000310_100 |
0.2841296926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004346_030 |
0.2844618867 |
- |
- |
sucrose phosphate phosphatase, putative [Ricinus communis] |
| 9 |
Hb_000827_060 |
0.2853454786 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 10 |
Hb_000422_120 |
0.2854053311 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1B [Jatropha curcas] |
| 11 |
Hb_069696_010 |
0.2881827386 |
- |
- |
hypothetical protein POPTR_0003s21190g [Populus trichocarpa] |
| 12 |
Hb_008725_170 |
0.2896672322 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Populus euphratica] |
| 13 |
Hb_000525_050 |
0.2903088925 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_181026_010 |
0.2916669828 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 15 |
Hb_006816_470 |
0.2919539496 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001008_180 |
0.2921787802 |
- |
- |
- |
| 17 |
Hb_001059_170 |
0.2928908477 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 18 |
Hb_012368_010 |
0.2935091031 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 19 |
Hb_000062_100 |
0.2980928635 |
- |
- |
- |
| 20 |
Hb_011386_020 |
0.2981076367 |
- |
- |
unnamed protein product [Vitis vinifera] |