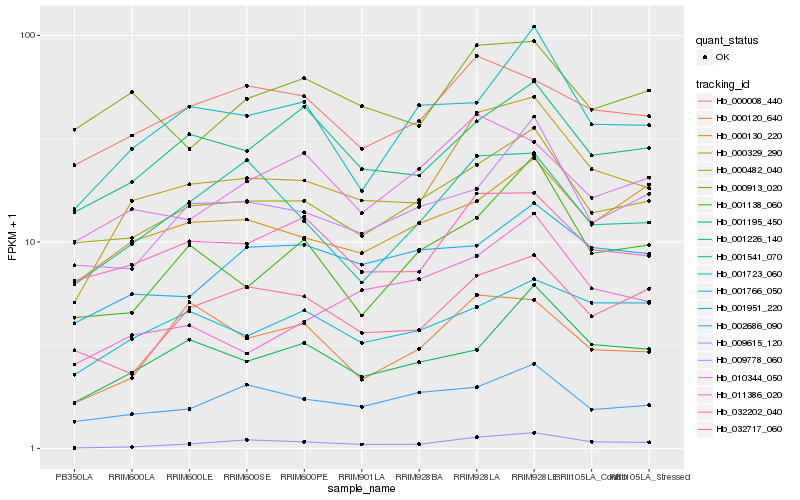
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_290 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 2 |
Hb_000482_040 |
0.1092574433 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 3 |
Hb_032202_040 |
0.1155755945 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |
| 4 |
Hb_009615_120 |
0.1230123167 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |
| 5 |
Hb_001195_450 |
0.1257935528 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 6 |
Hb_001226_140 |
0.1260460711 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 7 |
Hb_001723_060 |
0.1307503519 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 8 |
Hb_001541_070 |
0.1310452226 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 9 |
Hb_009778_060 |
0.1313851617 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 10 |
Hb_000008_440 |
0.1334038089 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Jatropha curcas] |
| 11 |
Hb_002686_090 |
0.1339829667 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 12 |
Hb_000130_220 |
0.1340007654 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_001138_060 |
0.1349572146 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 14 |
Hb_000120_640 |
0.1349855777 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_001766_050 |
0.1356214821 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29290 [Jatropha curcas] |
| 16 |
Hb_010344_050 |
0.1369728747 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 17 |
Hb_000913_020 |
0.1379232961 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 18 |
Hb_032717_060 |
0.13856154 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ18 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001951_220 |
0.1387301994 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 20 |
Hb_011386_020 |
0.1395927902 |
- |
- |
unnamed protein product [Vitis vinifera] |