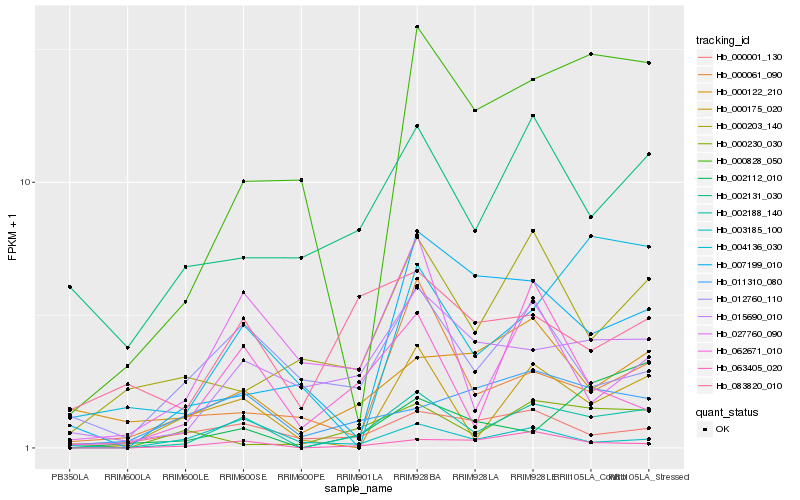
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002188_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650814 [Jatropha curcas] |
| 2 |
Hb_063405_020 |
0.2245620536 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 3 |
Hb_015690_010 |
0.2442036706 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g038521mg [Citrus sinensis] |
| 4 |
Hb_000175_020 |
0.2461878314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_062671_010 |
0.2761016015 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000828_050 |
0.2769218573 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000230_030 |
0.2778915301 |
- |
- |
hypothetical protein B456_013G2021001, partial [Gossypium raimondii] |
| 8 |
Hb_000061_090 |
0.2880773546 |
- |
- |
hypothetical protein B456_003G0981001, partial [Gossypium raimondii] |
| 9 |
Hb_002131_030 |
0.2883734184 |
- |
- |
PREDICTED: pyruvate kinase isozyme G, chloroplastic [Jatropha curcas] |
| 10 |
Hb_012760_110 |
0.2896718105 |
- |
- |
PREDICTED: methyltransferase-like protein 23 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000203_140 |
0.2903982404 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 12 |
Hb_011310_080 |
0.2907033752 |
- |
- |
hypothetical protein POPTR_0006s26840g [Populus trichocarpa] |
| 13 |
Hb_083820_010 |
0.2937385606 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 14 |
Hb_000001_130 |
0.2951979407 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 15 |
Hb_000122_210 |
0.2980904932 |
- |
- |
- |
| 16 |
Hb_004136_030 |
0.3019555666 |
- |
- |
- |
| 17 |
Hb_027760_090 |
0.3045424206 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_003185_100 |
0.3092314446 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 19 |
Hb_002112_010 |
0.3096035485 |
- |
- |
hypothetical protein POPTR_0001s45830g [Populus trichocarpa] |
| 20 |
Hb_007199_010 |
0.3106560429 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic [Nelumbo nucifera] |