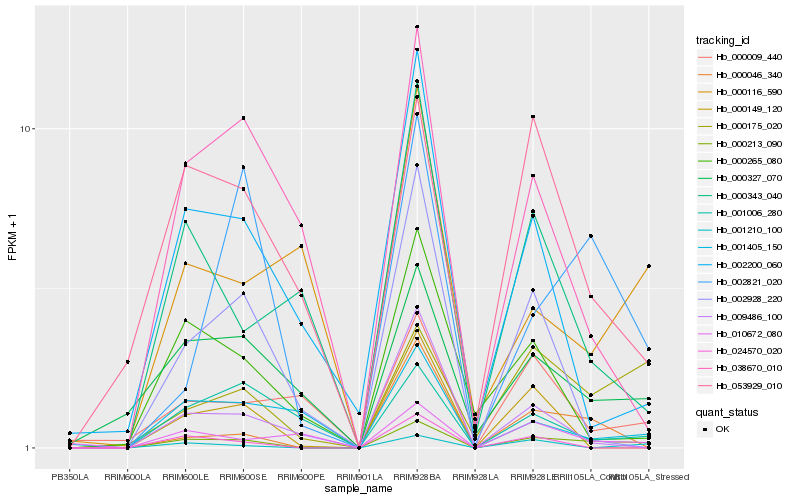
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000213_090 |
0.0 |
- |
- |
hypothetical protein OsI_39056 [Oryza sativa Indica Group] |
| 2 |
Hb_000175_020 |
0.229901392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001210_100 |
0.2602266191 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 4 |
Hb_000327_070 |
0.2628821756 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_053929_010 |
0.2744449477 |
- |
- |
hypothetical protein POPTR_0010s22320g [Populus trichocarpa] |
| 6 |
Hb_002821_020 |
0.2784990033 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter C family member 4 [Vitis vinifera] |
| 7 |
Hb_000046_340 |
0.2796687128 |
- |
- |
- |
| 8 |
Hb_000343_040 |
0.2946994336 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 9 |
Hb_010672_080 |
0.2994245421 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_00902 [Jatropha curcas] |
| 10 |
Hb_000009_440 |
0.3008282008 |
- |
- |
PREDICTED: uncharacterized protein LOC105639617 [Jatropha curcas] |
| 11 |
Hb_002200_060 |
0.3033518708 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 12 |
Hb_001006_280 |
0.3052005896 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 13 |
Hb_001405_150 |
0.3052991937 |
- |
- |
PREDICTED: protein At-4/1 [Jatropha curcas] |
| 14 |
Hb_000265_080 |
0.3058608896 |
- |
- |
Vacuolar cation/proton exchanger, putative [Ricinus communis] |
| 15 |
Hb_000116_590 |
0.3103236327 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 16 |
Hb_024570_020 |
0.3127305091 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 17 |
Hb_000149_120 |
0.3127750913 |
- |
- |
PREDICTED: uncharacterized protein LOC105642119 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002928_220 |
0.3128932893 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 19 |
Hb_038670_010 |
0.313020355 |
- |
- |
Major allergen Pru ar, putative [Ricinus communis] |
| 20 |
Hb_009486_100 |
0.3130328144 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |