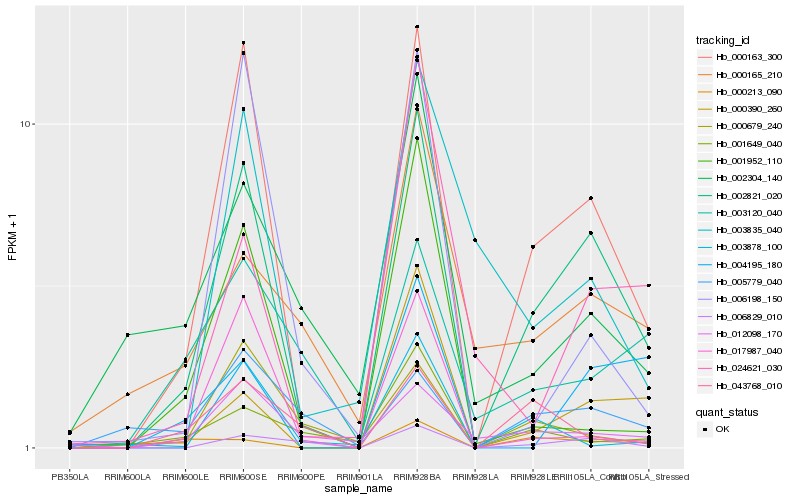
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002821_020 |
0.0 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter C family member 4 [Vitis vinifera] |
| 2 |
Hb_000163_300 |
0.0975495777 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 3 |
Hb_006829_010 |
0.2313558558 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000390_260 |
0.2637811678 |
transcription factor |
TF Family: MYB |
Myb domain protein 107 [Theobroma cacao] |
| 5 |
Hb_012098_170 |
0.2682985617 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_16874 [Jatropha curcas] |
| 6 |
Hb_000213_090 |
0.2784990033 |
- |
- |
hypothetical protein OsI_39056 [Oryza sativa Indica Group] |
| 7 |
Hb_017987_040 |
0.2862988643 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 8 |
Hb_024621_030 |
0.2888528245 |
- |
- |
tetraketide alpha-pyrone reductase 2 [Jatropha curcas] |
| 9 |
Hb_003835_040 |
0.2892914707 |
- |
- |
PREDICTED: UDP-glycosyltransferase 91A1-like [Jatropha curcas] |
| 10 |
Hb_000165_210 |
0.2914016241 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005779_040 |
0.2917297068 |
- |
- |
- |
| 12 |
Hb_043768_010 |
0.2962019798 |
- |
- |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 13 |
Hb_003878_100 |
0.2971363615 |
desease resistance |
Gene Name: NB-ARC |
LRR and NB-ARC domains-containing disease resistance protein, putative [Theobroma cacao] |
| 14 |
Hb_006198_150 |
0.2996680988 |
- |
- |
- |
| 15 |
Hb_002304_140 |
0.300773617 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_003120_040 |
0.3012211438 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 17 |
Hb_000679_240 |
0.3023923407 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 18 |
Hb_001649_040 |
0.3035118654 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 19 |
Hb_001952_110 |
0.3036489677 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_004195_180 |
0.3040663483 |
- |
- |
unnamed protein product [Coffea canephora] |