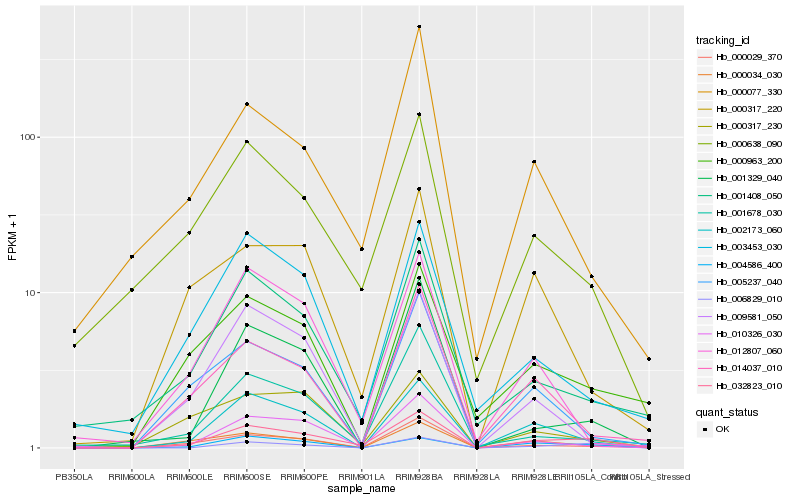
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032823_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002173_060 |
0.1793165507 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |
| 3 |
Hb_004586_400 |
0.192627905 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_000029_370 |
0.1950139836 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_000638_090 |
0.2000597362 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_010326_030 |
0.2046789214 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 4-like [Jatropha curcas] |
| 7 |
Hb_001408_050 |
0.2074760193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000963_200 |
0.2159375926 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 9 |
Hb_000317_220 |
0.2188802115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005237_040 |
0.2211703048 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 11 |
Hb_000317_230 |
0.2211939712 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 12 |
Hb_000077_330 |
0.2230970125 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2-like [Jatropha curcas] |
| 13 |
Hb_000034_030 |
0.2237890525 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_003453_030 |
0.2251130725 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 15 |
Hb_001329_040 |
0.2262497259 |
- |
- |
hypothetical protein RCOM_1345340 [Ricinus communis] |
| 16 |
Hb_001678_030 |
0.2264526008 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 17 |
Hb_006829_010 |
0.2265533173 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_012807_060 |
0.2268769955 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_014037_010 |
0.2270953506 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 20 |
Hb_009581_050 |
0.2287606436 |
- |
- |
hypothetical protein CISIN_1g0025812mg, partial [Citrus sinensis] |