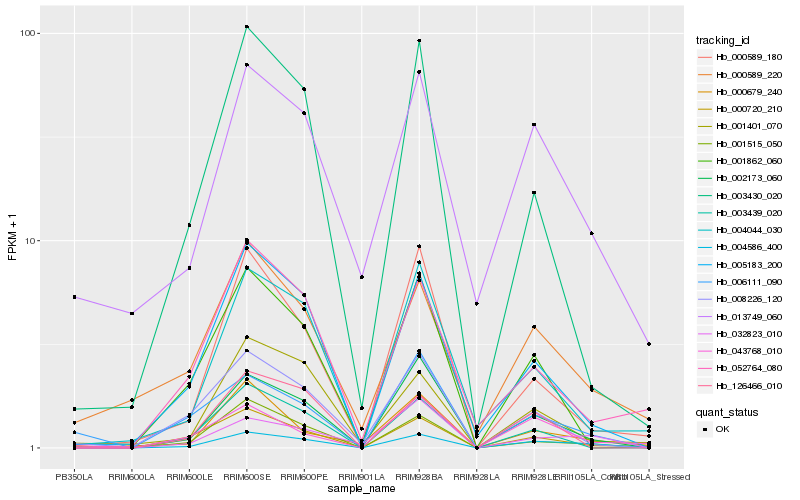
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004586_400 |
0.0 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_002173_060 |
0.1525910364 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |
| 3 |
Hb_001515_050 |
0.1794795788 |
- |
- |
PREDICTED: uncharacterized protein LOC105636626 [Jatropha curcas] |
| 4 |
Hb_005183_200 |
0.1851455879 |
- |
- |
EG964 [Manihot glaziovii] |
| 5 |
Hb_032823_010 |
0.192627905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000720_210 |
0.2005388482 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 7 |
Hb_043768_010 |
0.2145427183 |
- |
- |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 8 |
Hb_013749_060 |
0.2145648394 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 9 |
Hb_004044_030 |
0.2155236552 |
- |
- |
PREDICTED: uncharacterized protein LOC105639335 [Jatropha curcas] |
| 10 |
Hb_003439_020 |
0.2176968756 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1-like [Solanum lycopersicum] |
| 11 |
Hb_126466_010 |
0.220434593 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_008226_120 |
0.2215966369 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_001401_070 |
0.2217856554 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 14 |
Hb_003430_020 |
0.221814753 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_000589_220 |
0.222436107 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_001862_060 |
0.2225044803 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000589_180 |
0.2246507635 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 18 |
Hb_000679_240 |
0.2261347411 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 19 |
Hb_052764_080 |
0.2294012154 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006111_090 |
0.2311456336 |
- |
- |
hypothetical protein JCGZ_26825 [Jatropha curcas] |