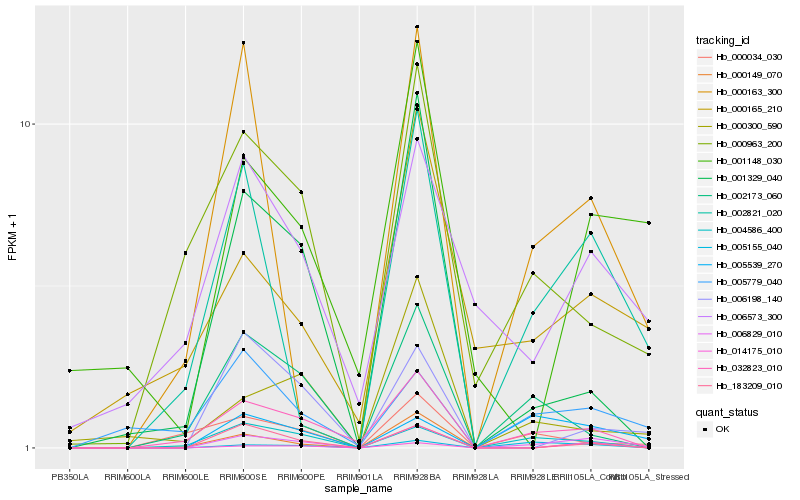
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006829_010 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_032823_010 |
0.2265533173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002821_020 |
0.2313558558 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter C family member 4 [Vitis vinifera] |
| 4 |
Hb_005539_270 |
0.261722464 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_004586_400 |
0.2646555446 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000163_300 |
0.2663273634 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 7 |
Hb_005155_040 |
0.2722574041 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 8 |
Hb_000149_070 |
0.2801431641 |
- |
- |
PREDICTED: uncharacterized protein At5g39570 isoform X1 [Jatropha curcas] |
| 9 |
Hb_183209_010 |
0.2808088163 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 10 |
Hb_014175_010 |
0.28467465 |
- |
- |
Uncharacterized protein TCM_033221 [Theobroma cacao] |
| 11 |
Hb_002173_060 |
0.2868548011 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |
| 12 |
Hb_005779_040 |
0.2870885227 |
- |
- |
- |
| 13 |
Hb_006198_140 |
0.2887116852 |
- |
- |
hypothetical protein L484_015208 [Morus notabilis] |
| 14 |
Hb_001329_040 |
0.2898840448 |
- |
- |
hypothetical protein RCOM_1345340 [Ricinus communis] |
| 15 |
Hb_000963_200 |
0.2912538106 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 16 |
Hb_000165_210 |
0.2945250769 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006573_300 |
0.2966855106 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_001148_030 |
0.3010694339 |
- |
- |
- |
| 19 |
Hb_000300_590 |
0.3018723666 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |
| 20 |
Hb_000034_030 |
0.3033597747 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |