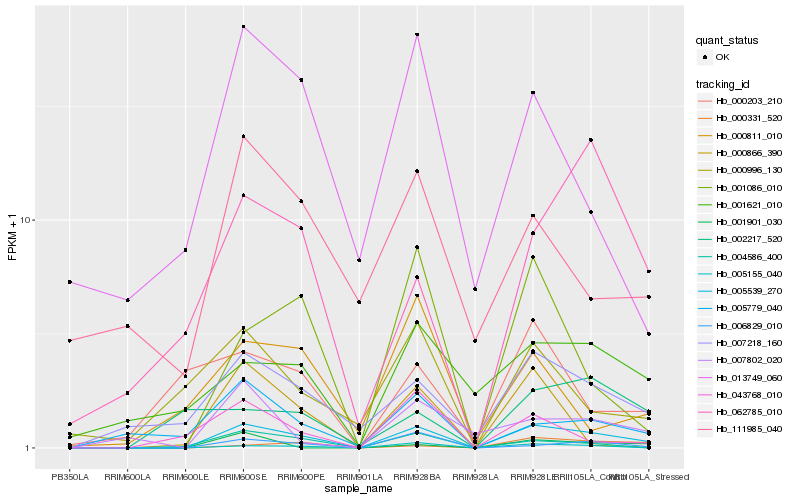
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_270 |
0.0 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_005155_040 |
0.2275389574 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 3 |
Hb_007218_160 |
0.253498403 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 4 |
Hb_006829_010 |
0.261722464 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_004586_400 |
0.2654332804 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_001086_010 |
0.2670963698 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 7 |
Hb_005779_040 |
0.281581934 |
- |
- |
- |
| 8 |
Hb_043768_010 |
0.2917383091 |
- |
- |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 9 |
Hb_001621_010 |
0.3031945313 |
- |
- |
PREDICTED: uncharacterized protein LOC102615432 [Citrus sinensis] |
| 10 |
Hb_007802_020 |
0.3064661022 |
- |
- |
PREDICTED: receptor-like protein 12 [Eucalyptus grandis] |
| 11 |
Hb_000331_520 |
0.3072152307 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 12 |
Hb_000866_390 |
0.3079289714 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 13 |
Hb_062785_010 |
0.3116568298 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 14 |
Hb_000996_130 |
0.3119636123 |
- |
- |
PREDICTED: remorin-like [Populus euphratica] |
| 15 |
Hb_013749_060 |
0.3153566818 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 16 |
Hb_000811_010 |
0.3217718751 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 17 |
Hb_001901_030 |
0.3227051356 |
- |
- |
- |
| 18 |
Hb_111985_040 |
0.3243314763 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 19 |
Hb_000203_210 |
0.3251410228 |
- |
- |
PREDICTED: uncharacterized protein LOC105040619 isoform X3 [Elaeis guineensis] |
| 20 |
Hb_002217_520 |
0.3257611626 |
- |
- |
PREDICTED: probable protein phosphatase 2C 42 isoform X3 [Populus euphratica] |