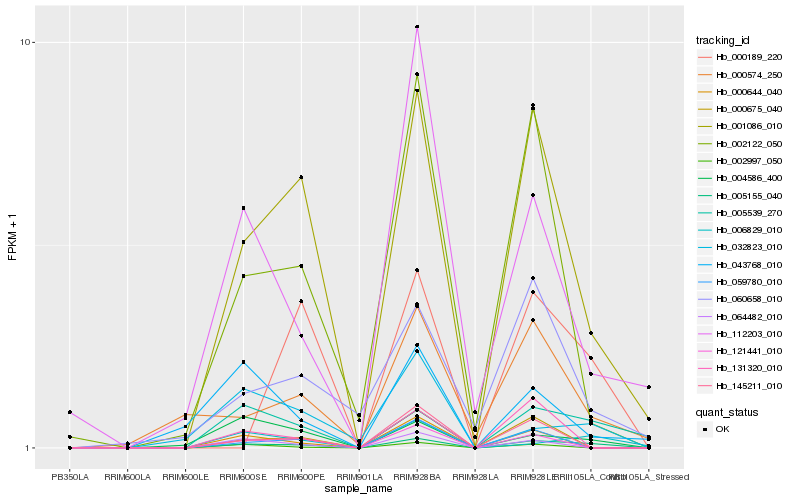
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005155_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 2 |
Hb_001086_010 |
0.1919062186 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 3 |
Hb_005539_270 |
0.2275389574 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_006829_010 |
0.2722574041 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_004586_400 |
0.272894662 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_002122_050 |
0.2830729748 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 7 |
Hb_000574_250 |
0.292164345 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 8 |
Hb_060658_010 |
0.2928555043 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 9 |
Hb_121441_010 |
0.2966708359 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_032823_010 |
0.3008757332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000189_220 |
0.3022127138 |
- |
- |
- |
| 12 |
Hb_000644_040 |
0.3038694937 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_009157 [Vitis vinifera] |
| 13 |
Hb_000675_040 |
0.3042213227 |
- |
- |
hypothetical protein OsI_02937 [Oryza sativa Indica Group] |
| 14 |
Hb_059780_010 |
0.3062103331 |
- |
- |
PREDICTED: uncharacterized protein LOC104885482, partial [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_112203_010 |
0.3064281318 |
- |
- |
- |
| 16 |
Hb_145211_010 |
0.3104910654 |
- |
- |
hypothetical protein M569_05648, partial [Genlisea aurea] |
| 17 |
Hb_064482_010 |
0.3128586076 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_131320_010 |
0.3151548424 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |
| 19 |
Hb_043768_010 |
0.316597637 |
- |
- |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 20 |
Hb_002997_050 |
0.3183732779 |
- |
- |
PREDICTED: receptor-like protein 12 isoform X4 [Populus euphratica] |