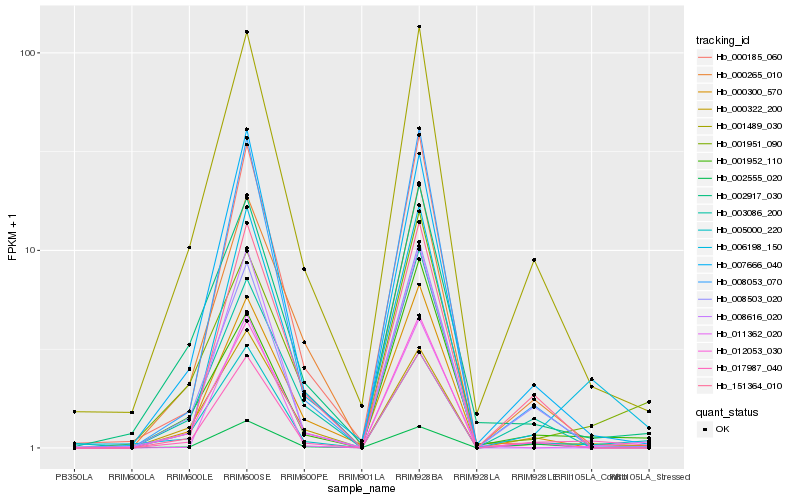
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017987_040 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 2 |
Hb_005000_220 |
0.0920083595 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 3 |
Hb_006198_150 |
0.1058200123 |
- |
- |
- |
| 4 |
Hb_001489_030 |
0.1174386845 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 5 |
Hb_008503_020 |
0.1180888197 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 6 |
Hb_007666_040 |
0.1198277182 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_008053_070 |
0.1273701771 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X4 [Jatropha curcas] |
| 8 |
Hb_012053_030 |
0.1282192751 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g22810 [Jatropha curcas] |
| 9 |
Hb_000300_570 |
0.1288223486 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 10 |
Hb_000322_200 |
0.1376817109 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 11 |
Hb_002555_020 |
0.144514073 |
- |
- |
Receptor-like protein kinase [Medicago truncatula] |
| 12 |
Hb_151364_010 |
0.1452319077 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 13 |
Hb_000265_010 |
0.1470217973 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_001952_110 |
0.1487936411 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000185_060 |
0.1515292096 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 16 |
Hb_008616_020 |
0.1537322221 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_002917_030 |
0.153914156 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Setaria italica] |
| 18 |
Hb_001951_090 |
0.1579470888 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 19 |
Hb_011362_020 |
0.1585023639 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_003086_200 |
0.1586489048 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |