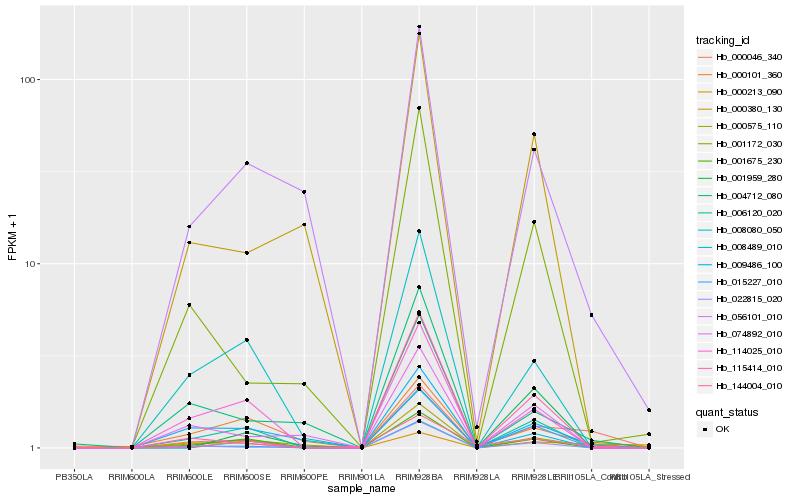
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_340 |
0.0 |
- |
- |
- |
| 2 |
Hb_000575_110 |
0.2402004226 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 3 |
Hb_009486_100 |
0.2633781223 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_144004_010 |
0.2776351232 |
- |
- |
hypothetical protein JCGZ_01009 [Jatropha curcas] |
| 5 |
Hb_000213_090 |
0.2796687128 |
- |
- |
hypothetical protein OsI_39056 [Oryza sativa Indica Group] |
| 6 |
Hb_008489_010 |
0.2809630497 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 7 |
Hb_001172_030 |
0.2824790418 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 7.2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001675_230 |
0.2860766125 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 9 |
Hb_115414_010 |
0.2866301307 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 10 |
Hb_006120_020 |
0.2870448731 |
- |
- |
- |
| 11 |
Hb_056101_010 |
0.2876609378 |
- |
- |
hypothetical protein POPTR_0391s00200g [Populus trichocarpa] |
| 12 |
Hb_114025_010 |
0.290173371 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 13 |
Hb_015227_010 |
0.2937215059 |
- |
- |
PREDICTED: uncharacterized protein LOC104903465 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_022815_020 |
0.2950993361 |
- |
- |
PREDICTED: uncharacterized protein LOC105650107 [Jatropha curcas] |
| 15 |
Hb_004712_080 |
0.2953618577 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 16 |
Hb_008080_050 |
0.2966892893 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_074892_010 |
0.2996441328 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 18 |
Hb_001959_280 |
0.3001989597 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 19 |
Hb_000101_360 |
0.3007192263 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 20 |
Hb_000380_130 |
0.3010383354 |
- |
- |
Phenylalanine ammonia-lyase, putative [Ricinus communis] |