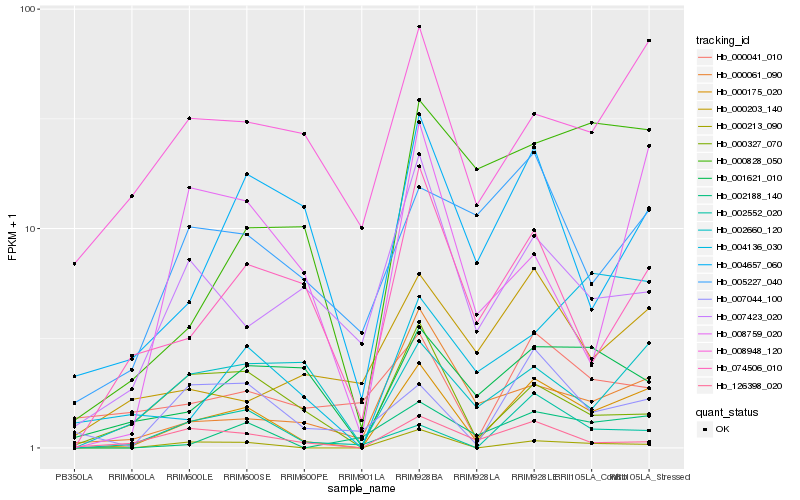
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000213_090 |
0.229901392 |
- |
- |
hypothetical protein OsI_39056 [Oryza sativa Indica Group] |
| 3 |
Hb_002188_140 |
0.2461878314 |
- |
- |
PREDICTED: uncharacterized protein LOC105650814 [Jatropha curcas] |
| 4 |
Hb_000061_090 |
0.2560923766 |
- |
- |
hypothetical protein B456_003G0981001, partial [Gossypium raimondii] |
| 5 |
Hb_074506_010 |
0.2602334892 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 6 |
Hb_004657_060 |
0.2715459738 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 7 |
Hb_007044_100 |
0.2776253589 |
- |
- |
hypothetical protein POPTR_0004s10420g [Populus trichocarpa] |
| 8 |
Hb_008948_120 |
0.2781901133 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000828_050 |
0.2808349266 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000327_070 |
0.2828250637 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_126398_020 |
0.2850504641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001621_010 |
0.2879267005 |
- |
- |
PREDICTED: uncharacterized protein LOC102615432 [Citrus sinensis] |
| 13 |
Hb_008759_020 |
0.2882164175 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 14 |
Hb_002660_120 |
0.2893788987 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Populus euphratica] |
| 15 |
Hb_000203_140 |
0.2915645943 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 16 |
Hb_002552_020 |
0.2923346712 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 17 |
Hb_007423_020 |
0.2934747275 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 18 |
Hb_004136_030 |
0.2934803509 |
- |
- |
- |
| 19 |
Hb_000041_010 |
0.2949252104 |
- |
- |
pfkB-type carbohydrate kinase family protein [Populus trichocarpa] |
| 20 |
Hb_005227_040 |
0.2953023531 |
transcription factor |
TF Family: HB |
unnamed protein product [Vitis vinifera] |