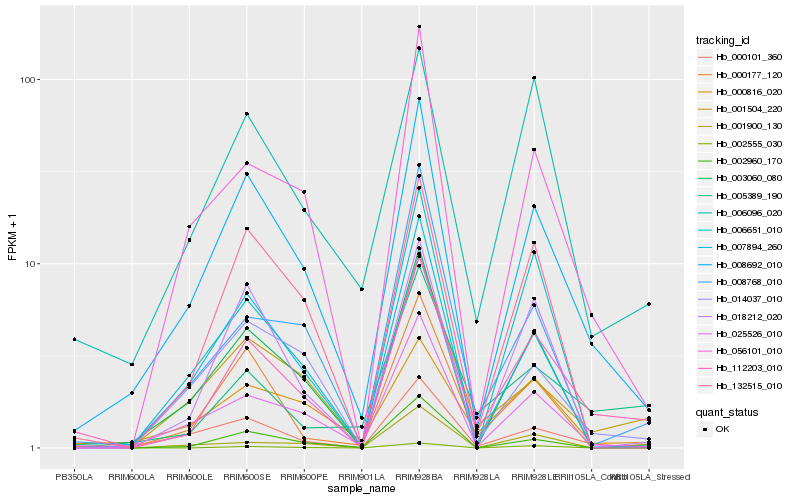
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_112203_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_008692_010 |
0.1620062898 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 3 |
Hb_001504_220 |
0.1843950262 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_005389_190 |
0.187359763 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 5 |
Hb_025526_010 |
0.1898374447 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 6 |
Hb_006096_020 |
0.1935046782 |
- |
- |
Os06g0343500 [Oryza sativa Japonica Group] |
| 7 |
Hb_056101_010 |
0.1962197973 |
- |
- |
hypothetical protein POPTR_0391s00200g [Populus trichocarpa] |
| 8 |
Hb_002960_170 |
0.2011999169 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 9 |
Hb_000101_360 |
0.203985631 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 10 |
Hb_132515_010 |
0.2063864096 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_000816_020 |
0.2074923028 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_001900_130 |
0.207883434 |
- |
- |
PREDICTED: uncharacterized protein LOC105632667 [Jatropha curcas] |
| 13 |
Hb_000177_120 |
0.209805968 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 14 |
Hb_018212_020 |
0.2112200023 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 15 |
Hb_008768_010 |
0.2119357593 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006651_010 |
0.2168145347 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4-like [Citrus sinensis] |
| 17 |
Hb_002555_030 |
0.2168835226 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Malus domestica] |
| 18 |
Hb_003060_080 |
0.2169803902 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_007894_260 |
0.2174693811 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_014037_010 |
0.219593621 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |