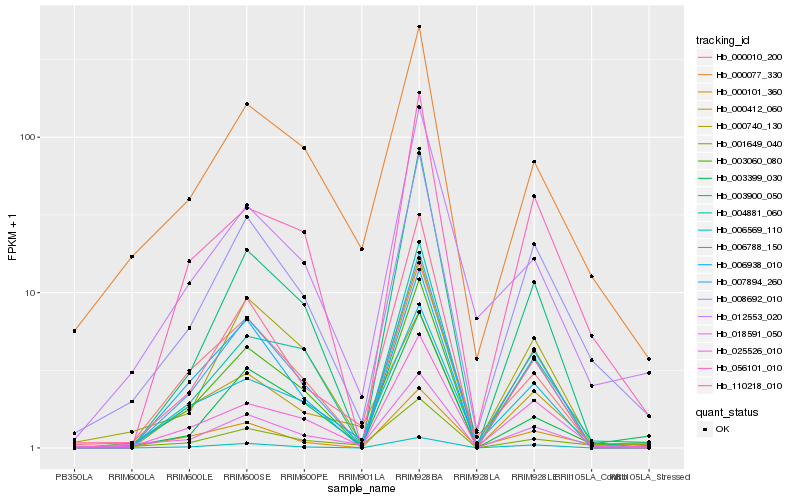
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018591_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 2 |
Hb_000077_330 |
0.1053407145 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2-like [Jatropha curcas] |
| 3 |
Hb_008692_010 |
0.109455842 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 4 |
Hb_025526_010 |
0.129619386 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 5 |
Hb_007894_260 |
0.133900645 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000412_060 |
0.1345574509 |
- |
- |
hypothetical protein JCGZ_18814 [Jatropha curcas] |
| 7 |
Hb_003060_080 |
0.1401353913 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 8 |
Hb_110218_010 |
0.1442858053 |
- |
- |
PREDICTED: uncharacterized protein LOC104428128 [Eucalyptus grandis] |
| 9 |
Hb_006788_150 |
0.1458516919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001649_040 |
0.1519301764 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 11 |
Hb_000740_130 |
0.1543883554 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 12 |
Hb_004881_060 |
0.1564147875 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 13 |
Hb_006569_110 |
0.158362536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000010_200 |
0.1588493295 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_012553_020 |
0.1591886103 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 16 |
Hb_000101_360 |
0.160424558 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 17 |
Hb_003900_050 |
0.1614063 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 18 |
Hb_006938_010 |
0.1620504573 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 19 |
Hb_003399_030 |
0.1633450288 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 20 |
Hb_056101_010 |
0.1636670891 |
- |
- |
hypothetical protein POPTR_0391s00200g [Populus trichocarpa] |