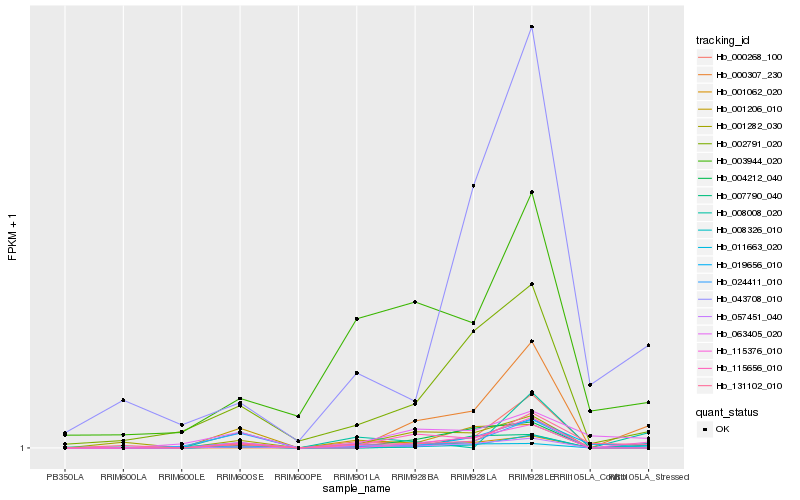
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001062_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103962405 [Pyrus x bretschneideri] |
| 2 |
Hb_115376_010 |
0.1724214459 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 3 |
Hb_002791_020 |
0.2454842938 |
- |
- |
hypothetical protein AALP_AA5G271700 [Arabis alpina] |
| 4 |
Hb_011663_020 |
0.2768038507 |
- |
- |
Putative retrotransposon protein, identical [Solanum demissum] |
| 5 |
Hb_057451_040 |
0.2775183236 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_001206_010 |
0.2858409965 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 7 |
Hb_063405_020 |
0.2926167999 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 8 |
Hb_115656_010 |
0.2962314595 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 9 |
Hb_131102_010 |
0.2997282141 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_024411_010 |
0.3033952028 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_007790_040 |
0.3038184357 |
- |
- |
PREDICTED: uncharacterized protein LOC104417177 [Eucalyptus grandis] |
| 12 |
Hb_019656_010 |
0.3046694765 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_043708_010 |
0.3059228107 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 14 |
Hb_008326_010 |
0.3073661347 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 15 |
Hb_004212_040 |
0.3234253011 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_003944_020 |
0.3279796943 |
- |
- |
PREDICTED: uncharacterized protein LOC105110580 [Populus euphratica] |
| 17 |
Hb_001282_030 |
0.3280707148 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 18 |
Hb_000307_230 |
0.3307603439 |
- |
- |
- |
| 19 |
Hb_000268_100 |
0.3326191219 |
- |
- |
- |
| 20 |
Hb_008008_020 |
0.334372641 |
- |
- |
- |