| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
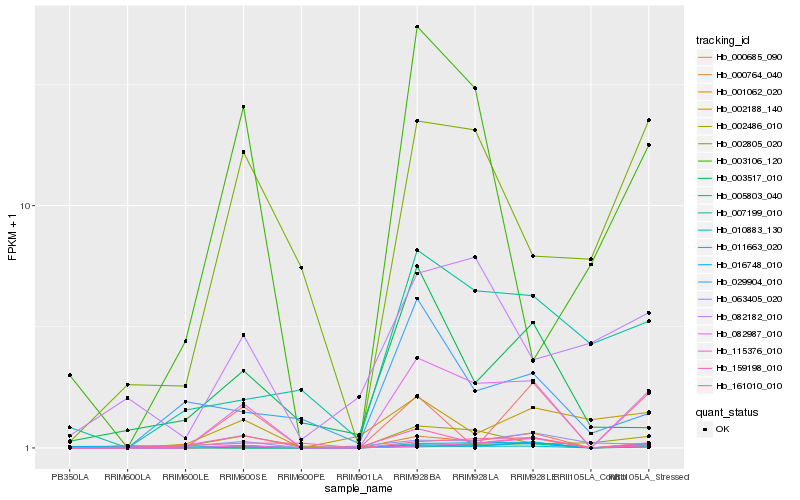
Hb_082987_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10030423mg [Citrus clementina] |
| 2 |
Hb_011663_020 |
0.2761750202 |
- |
- |
Putative retrotransposon protein, identical [Solanum demissum] |
| 3 |
Hb_010883_130 |
0.2800973176 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210 [Jatropha curcas] |
| 4 |
Hb_115376_010 |
0.3005936484 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 5 |
Hb_000764_040 |
0.3007827732 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 6 |
Hb_159198_010 |
0.313113175 |
- |
- |
hypothetical protein VITISV_033829 [Vitis vinifera] |
| 7 |
Hb_002805_020 |
0.322959057 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_002486_010 |
0.3236104186 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 1 [Jatropha curcas] |
| 9 |
Hb_161010_010 |
0.3270094918 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 10 |
Hb_007199_010 |
0.3295963182 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic [Nelumbo nucifera] |
| 11 |
Hb_001062_020 |
0.3385906647 |
- |
- |
PREDICTED: uncharacterized protein LOC103962405 [Pyrus x bretschneideri] |
| 12 |
Hb_003106_120 |
0.3449136977 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 13 |
Hb_005803_040 |
0.3461974886 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_000685_090 |
0.3486357761 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 15 |
Hb_082182_010 |
0.3513999436 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 16 |
Hb_016748_010 |
0.3540140613 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_002188_140 |
0.3545954896 |
- |
- |
PREDICTED: uncharacterized protein LOC105650814 [Jatropha curcas] |
| 18 |
Hb_063405_020 |
0.3569206794 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 19 |
Hb_029904_010 |
0.3595756021 |
- |
- |
PREDICTED: uncharacterized protein LOC105635075 [Jatropha curcas] |
| 20 |
Hb_003517_010 |
0.3610627525 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |