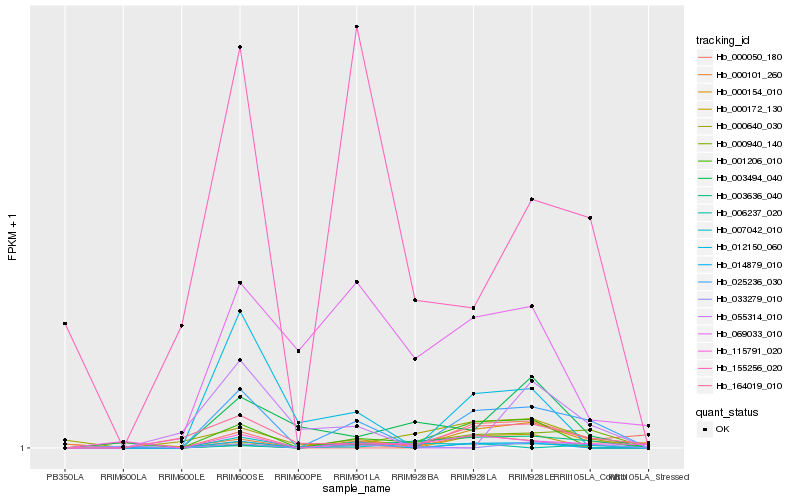
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025236_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 2 |
Hb_014879_010 |
0.2809563563 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_001206_010 |
0.341641031 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 4 |
Hb_164019_010 |
0.3480367047 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_012150_060 |
0.3481525963 |
- |
- |
- |
| 6 |
Hb_155256_020 |
0.3731497246 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 7 |
Hb_000940_140 |
0.380201955 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 8 |
Hb_115791_020 |
0.3816653059 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 9 |
Hb_003636_040 |
0.3929501213 |
- |
- |
mitochondrial translational initiation factor, putative [Ricinus communis] |
| 10 |
Hb_007042_010 |
0.397161343 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_000172_130 |
0.4047703251 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 12 |
Hb_069033_010 |
0.4055409363 |
desease resistance |
Gene Name: DUF720 |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 13 |
Hb_000154_010 |
0.4079910098 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_000101_260 |
0.4086119311 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_055314_010 |
0.4231695955 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 16 |
Hb_000640_030 |
0.423315017 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 17 |
Hb_003494_040 |
0.4263477488 |
transcription factor |
TF Family: NAC |
PREDICTED: protein FEZ [Jatropha curcas] |
| 18 |
Hb_006237_020 |
0.4279856737 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 19 |
Hb_000050_180 |
0.4309241371 |
- |
- |
- |
| 20 |
Hb_033279_010 |
0.4310979461 |
- |
- |
DNA/RNA polymerases superfamily protein, putative [Theobroma cacao] |