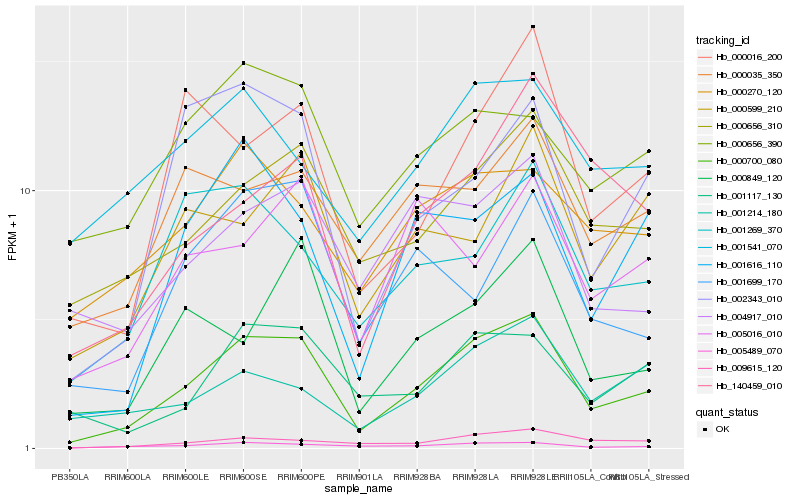
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000700_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005489_070 |
0.1597036906 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 3 |
Hb_000656_310 |
0.1598806247 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 4 |
Hb_009615_120 |
0.1643997717 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |
| 5 |
Hb_001117_130 |
0.1689134078 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 6 |
Hb_001616_110 |
0.1716943237 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 7 |
Hb_002343_010 |
0.1726475939 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_000849_120 |
0.1741869388 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 9 |
Hb_000016_200 |
0.1776442465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000270_120 |
0.1818897905 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 11 |
Hb_004917_010 |
0.183345029 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 12 |
Hb_001699_170 |
0.1840379102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000035_350 |
0.184326534 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000656_390 |
0.1850654588 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000599_210 |
0.1851256663 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 16 |
Hb_001269_370 |
0.1862957471 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 17 |
Hb_001214_180 |
0.1885067055 |
- |
- |
- |
| 18 |
Hb_140459_010 |
0.1895417571 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 19 |
Hb_005016_010 |
0.1913833915 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 20 |
Hb_001541_070 |
0.1920993042 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |