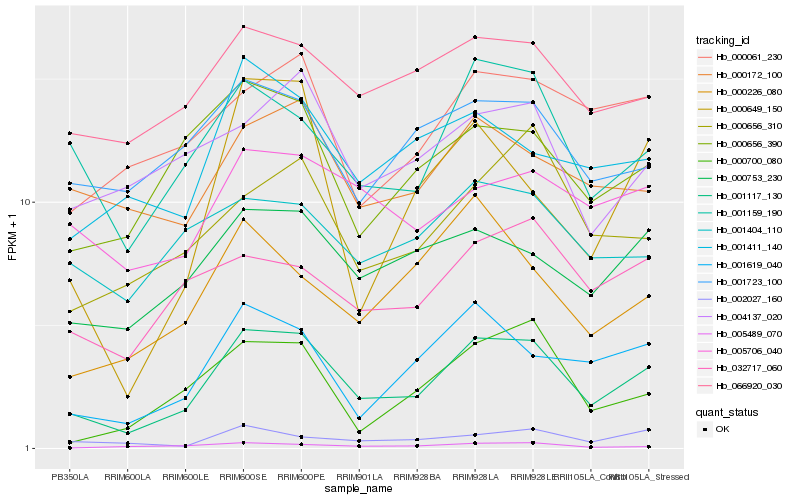
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001117_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 2 |
Hb_001159_190 |
0.1425072737 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_001619_040 |
0.1454189045 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 4 |
Hb_000753_230 |
0.149608098 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000656_390 |
0.1572825695 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000656_310 |
0.1573877824 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 7 |
Hb_066920_030 |
0.1606831102 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 8 |
Hb_032717_060 |
0.1621176412 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ18 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000649_150 |
0.1625668583 |
- |
- |
PREDICTED: uncharacterized protein LOC105634115 [Jatropha curcas] |
| 10 |
Hb_002027_160 |
0.1650980722 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_000061_230 |
0.1663163571 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 12 |
Hb_001411_140 |
0.167735016 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 13 |
Hb_000172_100 |
0.1683729173 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 2, peroxisomal [Jatropha curcas] |
| 14 |
Hb_000226_080 |
0.1684153877 |
- |
- |
prephenate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000700_080 |
0.1689134078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005706_040 |
0.1694270172 |
- |
- |
PREDICTED: uncharacterized protein LOC105642418 [Jatropha curcas] |
| 17 |
Hb_005489_070 |
0.1700936385 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 18 |
Hb_001723_100 |
0.1707790988 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004137_020 |
0.1721296464 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001404_110 |
0.1723584713 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |