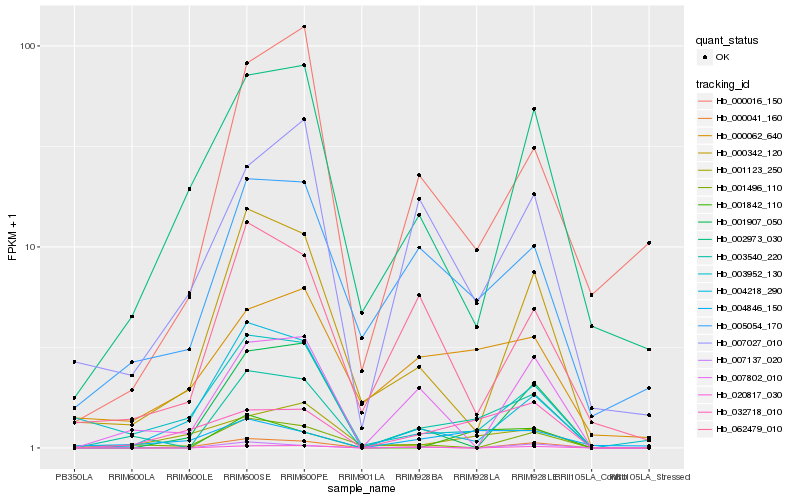
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003540_220 |
0.0 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 2 |
Hb_005054_170 |
0.254648263 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_007802_010 |
0.2605513815 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 4 |
Hb_032718_010 |
0.2625928265 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 5 |
Hb_000342_120 |
0.2673294674 |
- |
- |
PREDICTED: probable protein phosphatase 2C 72 [Jatropha curcas] |
| 6 |
Hb_001842_110 |
0.2704780509 |
transcription factor |
TF Family: ERF |
CBF-like transcription factor, putative [Ricinus communis] |
| 7 |
Hb_020817_030 |
0.2732824171 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 8 |
Hb_001496_110 |
0.2762172474 |
- |
- |
hypothetical protein VITISV_044023 [Vitis vinifera] |
| 9 |
Hb_007027_010 |
0.2789107752 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 10 |
Hb_062479_010 |
0.2837475575 |
- |
- |
hypothetical protein PHAVU_008G071200g [Phaseolus vulgaris] |
| 11 |
Hb_004218_290 |
0.286250207 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 6 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001907_050 |
0.2873194176 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 13 |
Hb_000062_640 |
0.2875244988 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 14 |
Hb_000016_150 |
0.2892497862 |
- |
- |
PREDICTED: uncharacterized protein LOC105137629 [Populus euphratica] |
| 15 |
Hb_004846_150 |
0.2941832016 |
- |
- |
hypothetical protein JCGZ_26968 [Jatropha curcas] |
| 16 |
Hb_001123_250 |
0.2942960558 |
- |
- |
hypothetical protein POPTR_0001s13220g [Populus trichocarpa] |
| 17 |
Hb_003952_130 |
0.2951976316 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 18 |
Hb_007137_020 |
0.2952877277 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 19 |
Hb_002973_030 |
0.2961123129 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000041_160 |
0.2985951136 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |