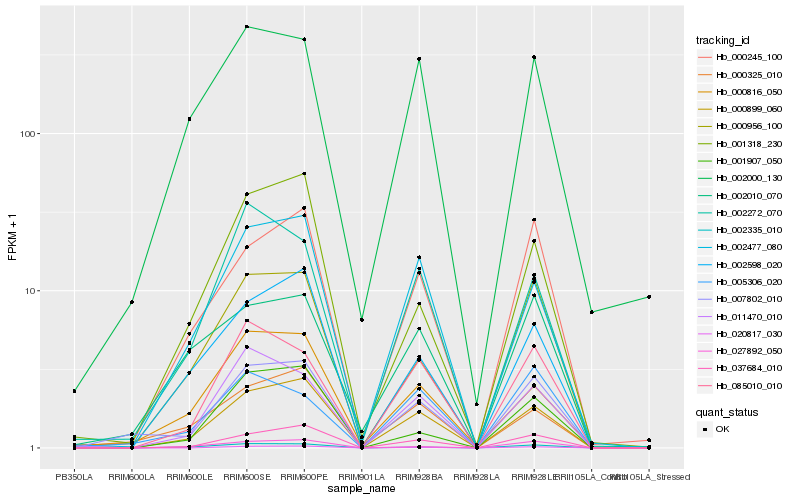
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007802_010 |
0.0 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 2 |
Hb_000899_060 |
0.1382827031 |
- |
- |
PREDICTED: uncharacterized protein LOC105793631 [Gossypium raimondii] |
| 3 |
Hb_037684_010 |
0.1454647283 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 4 |
Hb_000956_100 |
0.158506563 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 5 |
Hb_020817_030 |
0.161537185 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 6 |
Hb_000325_010 |
0.1644532885 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 7 |
Hb_000245_100 |
0.1647506923 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 8 |
Hb_085010_010 |
0.1729938652 |
- |
- |
PREDICTED: MLO-like protein 3 isoform X2 [Vitis vinifera] |
| 9 |
Hb_002335_010 |
0.1738569396 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 10 |
Hb_000816_050 |
0.1738754652 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 11 |
Hb_011470_010 |
0.1742392415 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 12 |
Hb_027892_050 |
0.1764665649 |
- |
- |
hypothetical protein PHAVU_009G226400g [Phaseolus vulgaris] |
| 13 |
Hb_002477_080 |
0.1818473322 |
- |
- |
hypothetical protein RCOM_0653450 [Ricinus communis] |
| 14 |
Hb_002598_020 |
0.1825775666 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |
| 15 |
Hb_002000_130 |
0.183829452 |
- |
- |
PREDICTED: abscisic stress-ripening protein 2-like [Eucalyptus grandis] |
| 16 |
Hb_001907_050 |
0.1851071875 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 17 |
Hb_002272_070 |
0.1883265348 |
- |
- |
PREDICTED: keratin, type I cytoskeletal 9 [Jatropha curcas] |
| 18 |
Hb_002010_070 |
0.1891531714 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 19 |
Hb_001318_230 |
0.1892526243 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 20 |
Hb_005306_020 |
0.1892871967 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |