| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
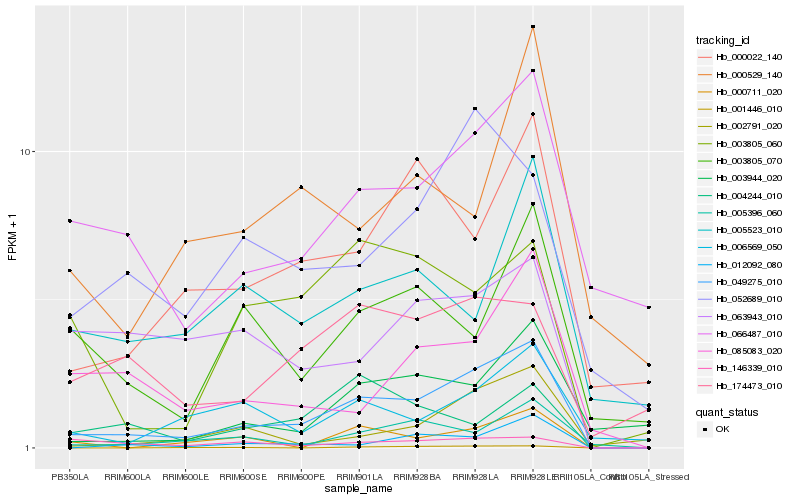
Hb_049275_010 |
0.0 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4-like [Populus euphratica] |
| 2 |
Hb_005396_060 |
0.227973885 |
- |
- |
- |
| 3 |
Hb_012092_080 |
0.231695367 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280-like [Malus domestica] |
| 4 |
Hb_000022_140 |
0.2349044422 |
- |
- |
hypothetical protein MIMGU_mgv1a009920mg [Erythranthe guttata] |
| 5 |
Hb_066487_010 |
0.237209767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006569_050 |
0.2404055857 |
- |
- |
- |
| 7 |
Hb_003944_020 |
0.2413688107 |
- |
- |
PREDICTED: uncharacterized protein LOC105110580 [Populus euphratica] |
| 8 |
Hb_052689_010 |
0.2453267935 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 9 |
Hb_085083_020 |
0.2460444988 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 10 |
Hb_000711_020 |
0.2471465045 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 11 |
Hb_002791_020 |
0.2478025122 |
- |
- |
hypothetical protein AALP_AA5G271700 [Arabis alpina] |
| 12 |
Hb_174473_010 |
0.2528881628 |
- |
- |
PREDICTED: uncharacterized protein LOC105775505 [Gossypium raimondii] |
| 13 |
Hb_003805_070 |
0.2567868103 |
- |
- |
PREDICTED: sacsin [Jatropha curcas] |
| 14 |
Hb_005523_010 |
0.2646269559 |
transcription factor |
TF Family: PHD |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 15 |
Hb_000529_140 |
0.2658911933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003805_060 |
0.2677237043 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_004244_010 |
0.2682035383 |
- |
- |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 18 |
Hb_146339_010 |
0.2688278684 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 19 |
Hb_063943_010 |
0.2689483842 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 20 |
Hb_001446_010 |
0.2705803537 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |