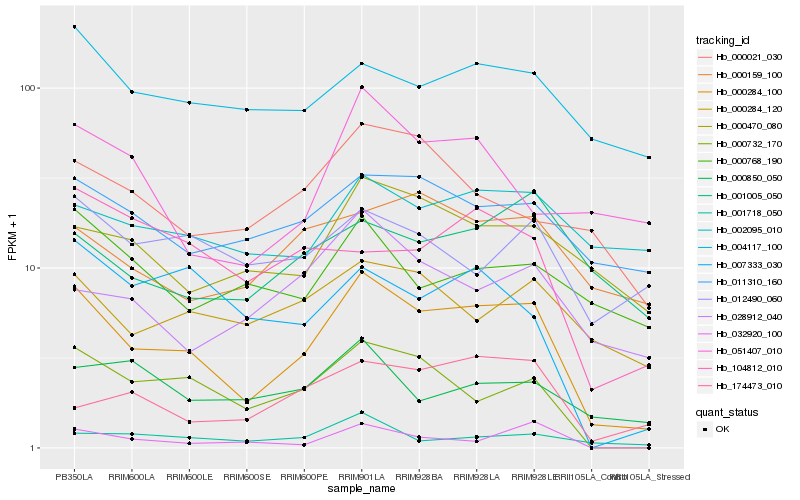
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000284_100 |
0.0 |
- |
- |
PREDICTED: transcription factor GTE11 isoform X2 [Populus euphratica] |
| 2 |
Hb_000732_170 |
0.1785444 |
- |
- |
PREDICTED: putative PAP-specific phosphatase, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_011310_160 |
0.2006059307 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 4 |
Hb_000470_080 |
0.2019432941 |
- |
- |
Caldesmon, putative [Ricinus communis] |
| 5 |
Hb_004117_100 |
0.203979154 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000284_120 |
0.2039971764 |
- |
- |
PREDICTED: uncharacterized protein LOC105629656 [Jatropha curcas] |
| 7 |
Hb_028912_040 |
0.2100582795 |
- |
- |
hypothetical protein POPTR_0016s03250g [Populus trichocarpa] |
| 8 |
Hb_000021_030 |
0.2117464107 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 9 |
Hb_001718_050 |
0.2129831674 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Brassica rapa] |
| 10 |
Hb_174473_010 |
0.2135106058 |
- |
- |
PREDICTED: uncharacterized protein LOC105775505 [Gossypium raimondii] |
| 11 |
Hb_032920_100 |
0.2136739262 |
- |
- |
hypothetical protein POPTR_0002s00940g, partial [Populus trichocarpa] |
| 12 |
Hb_007333_030 |
0.213827412 |
- |
- |
- |
| 13 |
Hb_000159_100 |
0.214087893 |
- |
- |
PREDICTED: uncharacterized protein At4g19900 isoform X2 [Jatropha curcas] |
| 14 |
Hb_104812_010 |
0.216066051 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 9 [Populus euphratica] |
| 15 |
Hb_000850_050 |
0.2172923153 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 16 |
Hb_051407_010 |
0.2182639504 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 17 |
Hb_012490_060 |
0.2186130228 |
- |
- |
PREDICTED: UBP1-associated proteins 1C isoform X1 [Jatropha curcas] |
| 18 |
Hb_001005_050 |
0.2193373887 |
- |
- |
hypothetical protein Csa_3G599480 [Cucumis sativus] |
| 19 |
Hb_000768_190 |
0.21992163 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 20 |
Hb_002095_010 |
0.2206345858 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |