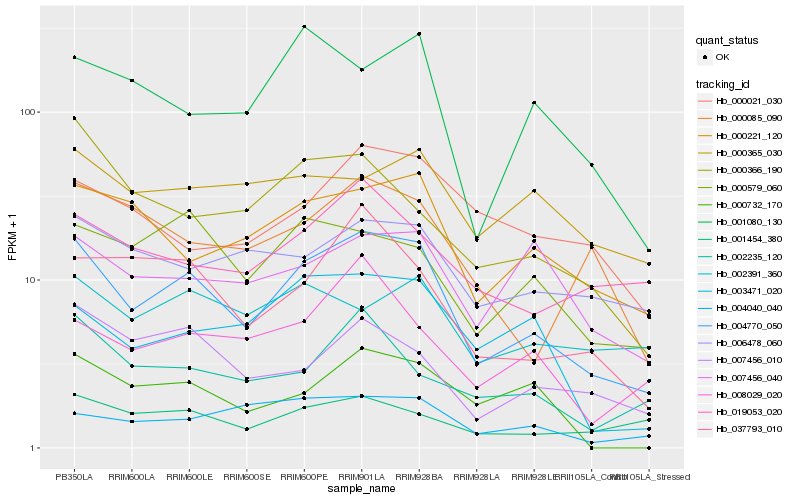
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004770_050 |
0.0 |
- |
- |
PREDICTED: serine carboxypeptidase-like 35 [Populus euphratica] |
| 2 |
Hb_000221_120 |
0.1622402411 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 3 |
Hb_037793_010 |
0.1674059061 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like [Jatropha curcas] |
| 4 |
Hb_000579_060 |
0.1679790309 |
- |
- |
PREDICTED: uncharacterized protein LOC105633849 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007456_010 |
0.1759084586 |
- |
- |
hypothetical protein L484_003540 [Morus notabilis] |
| 6 |
Hb_003471_020 |
0.1779995914 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8A [Jatropha curcas] |
| 7 |
Hb_006478_060 |
0.1807317954 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_019053_020 |
0.1807846838 |
- |
- |
hypothetical protein MIMGU_mgv1a0105441mg, partial [Erythranthe guttata] |
| 9 |
Hb_001080_130 |
0.1832307016 |
- |
- |
catalase [Hevea brasiliensis] |
| 10 |
Hb_008029_020 |
0.1843059741 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 11 |
Hb_000366_190 |
0.1844440793 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 12 |
Hb_004040_040 |
0.1848107833 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 13 |
Hb_007456_040 |
0.1851468346 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 14 |
Hb_001454_380 |
0.1871387535 |
- |
- |
PREDICTED: Fanconi anemia group I protein [Jatropha curcas] |
| 15 |
Hb_002235_120 |
0.1871413846 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 16 |
Hb_000085_090 |
0.1877121336 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 17 |
Hb_002391_360 |
0.1892775645 |
- |
- |
hypothetical protein CICLE_v10026208mg [Citrus clementina] |
| 18 |
Hb_000021_030 |
0.1918485532 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 19 |
Hb_000732_170 |
0.1922580775 |
- |
- |
PREDICTED: putative PAP-specific phosphatase, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_000365_030 |
0.1927565481 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |