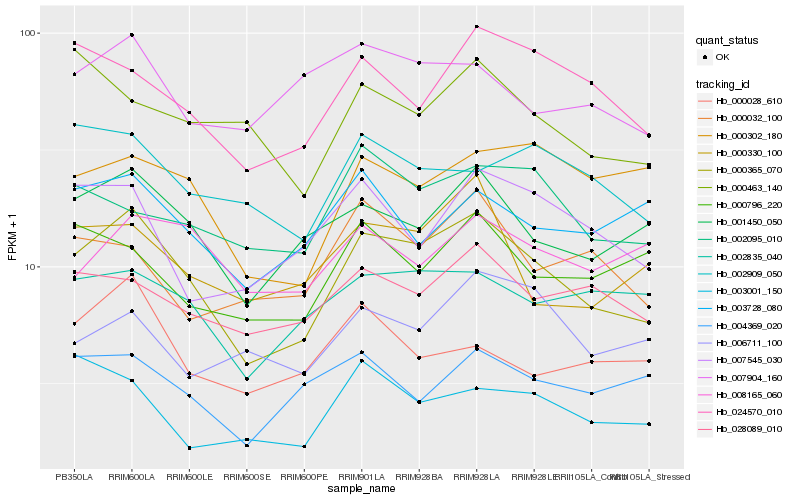
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_070 |
0.0 |
- |
- |
PREDICTED: SWR1 complex subunit 2 [Jatropha curcas] |
| 2 |
Hb_001450_050 |
0.1138290072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_024570_010 |
0.129565545 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 4 |
Hb_028089_010 |
0.1334966408 |
- |
- |
PREDICTED: homoserine dehydrogenase [Jatropha curcas] |
| 5 |
Hb_000330_100 |
0.1335942353 |
- |
- |
hypothetical protein VITISV_011650 [Vitis vinifera] |
| 6 |
Hb_002835_040 |
0.1337247284 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_007545_030 |
0.1392855741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002095_010 |
0.1398816596 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002909_050 |
0.1436735514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007904_160 |
0.1437038592 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 11 |
Hb_000302_180 |
0.1444678981 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000032_100 |
0.1448230554 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Vitis vinifera] |
| 13 |
Hb_000796_220 |
0.1459679416 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X3 [Jatropha curcas] |
| 14 |
Hb_003728_080 |
0.1462077784 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3-like [Jatropha curcas] |
| 15 |
Hb_004369_020 |
0.1466967515 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 16 |
Hb_000028_610 |
0.1494123873 |
- |
- |
PREDICTED: uncharacterized protein LOC105644291 [Jatropha curcas] |
| 17 |
Hb_008165_060 |
0.1500449829 |
- |
- |
PREDICTED: uncharacterized protein LOC105635541 [Jatropha curcas] |
| 18 |
Hb_003001_150 |
0.1508886775 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_006711_100 |
0.15172899 |
- |
- |
PREDICTED: uncharacterized protein LOC105637078 [Jatropha curcas] |
| 20 |
Hb_000463_140 |
0.1519234302 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |