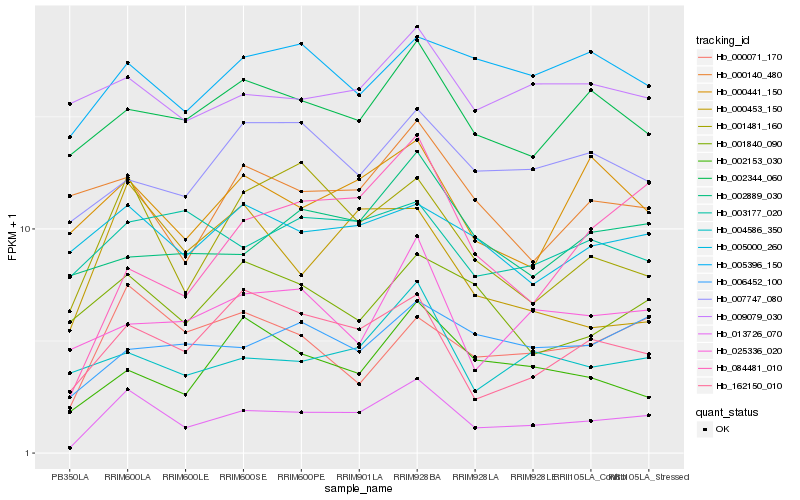
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013726_070 |
0.0 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Jatropha curcas] |
| 2 |
Hb_001481_160 |
0.1615187101 |
- |
- |
PREDICTED: RING-H2 finger protein ATL22-like [Populus euphratica] |
| 3 |
Hb_162150_010 |
0.1616635344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000071_170 |
0.1648494159 |
- |
- |
PREDICTED: uncharacterized protein LOC105628261 [Jatropha curcas] |
| 5 |
Hb_002344_060 |
0.1705026256 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 6 |
Hb_004586_350 |
0.1709817114 |
- |
- |
Phosphoribosylaminoimidazole-succinocarboxamide synthase [Morus notabilis] |
| 7 |
Hb_000441_150 |
0.1776493596 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_006452_100 |
0.1814391932 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 9 |
Hb_005000_260 |
0.1847552333 |
- |
- |
recA family protein [Populus trichocarpa] |
| 10 |
Hb_009079_030 |
0.1854924164 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 11 |
Hb_025336_020 |
0.1858782132 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 12 |
Hb_084481_010 |
0.1864127413 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 13 |
Hb_002153_030 |
0.186863707 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 14 |
Hb_002889_030 |
0.1875346385 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 [Jatropha curcas] |
| 15 |
Hb_005396_150 |
0.1877766306 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_000453_150 |
0.1886798364 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 17 |
Hb_007747_080 |
0.1890415492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001840_090 |
0.1892080702 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 19 |
Hb_000140_480 |
0.1893899792 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 20 |
Hb_003177_020 |
0.1911206616 |
- |
- |
diphthine synthase, putative [Ricinus communis] |