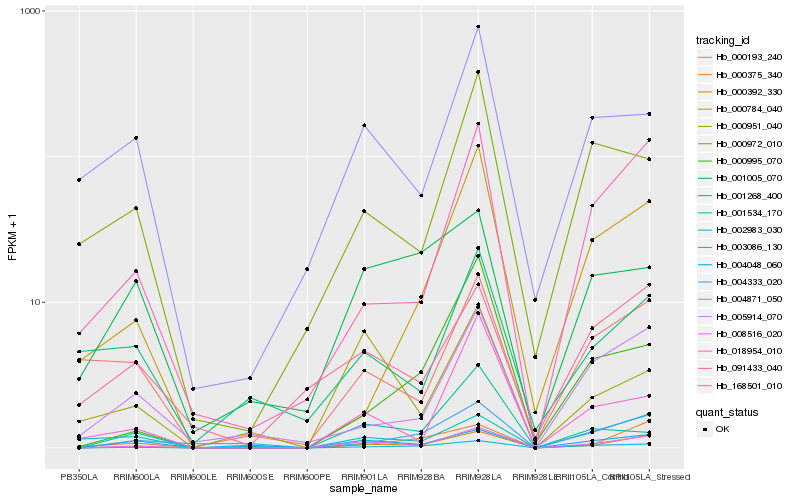
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008516_020 |
0.0 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |
| 2 |
Hb_000392_330 |
0.1424934808 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_003086_130 |
0.1565953892 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 4 |
Hb_004048_060 |
0.216035179 |
- |
- |
hypothetical protein PRUPE_ppa023788mg, partial [Prunus persica] |
| 5 |
Hb_091433_040 |
0.2223216157 |
- |
- |
PREDICTED: josephin-like protein [Jatropha curcas] |
| 6 |
Hb_001534_170 |
0.2386071739 |
- |
- |
Uncharacterized protein TCM_038456 [Theobroma cacao] |
| 7 |
Hb_000951_040 |
0.2409832444 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_09931 [Jatropha curcas] |
| 8 |
Hb_018954_010 |
0.2427542177 |
- |
- |
- |
| 9 |
Hb_004333_020 |
0.2429404262 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 10 |
Hb_004871_050 |
0.2437333064 |
- |
- |
hypothetical protein F383_22684 [Gossypium arboreum] |
| 11 |
Hb_005914_070 |
0.2470635403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000375_340 |
0.2507788741 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Populus euphratica] |
| 13 |
Hb_168501_010 |
0.2517439659 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000995_070 |
0.2521653422 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 15 |
Hb_000972_010 |
0.2548257778 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 16 |
Hb_000193_240 |
0.256728837 |
- |
- |
hypothetical protein CISIN_1g0449881mg, partial [Citrus sinensis] |
| 17 |
Hb_002983_030 |
0.2574685803 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g046124mg, partial [Citrus sinensis] |
| 18 |
Hb_000784_040 |
0.2642231713 |
- |
- |
hypothetical protein POPTR_0013s11550g [Populus trichocarpa] |
| 19 |
Hb_001268_400 |
0.2703245095 |
- |
- |
PREDICTED: uncharacterized protein LOC104583970 [Brachypodium distachyon] |
| 20 |
Hb_001005_070 |
0.276141102 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |