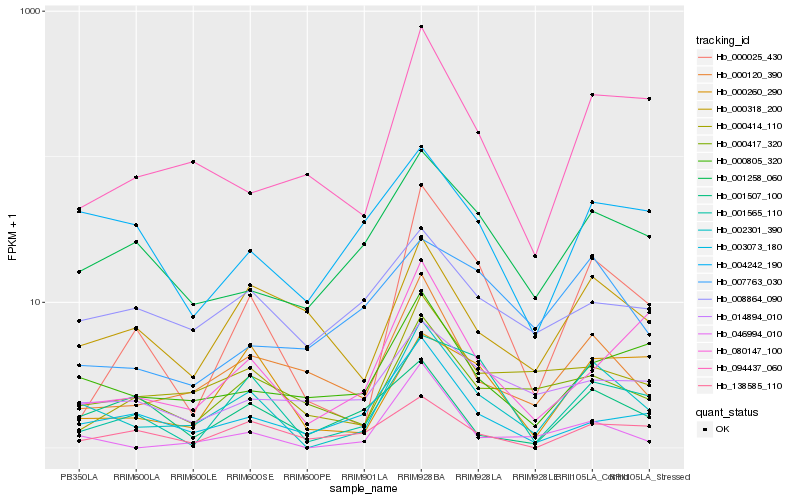
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_390 |
0.0 |
- |
- |
- |
| 2 |
Hb_000025_430 |
0.2239181619 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_000120_390 |
0.2248369415 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 4 |
Hb_046994_010 |
0.2294255377 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000417_320 |
0.2427500464 |
- |
- |
hypothetical protein CICLE_v10000333mg [Citrus clementina] |
| 6 |
Hb_094437_060 |
0.2451164512 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |
| 7 |
Hb_000805_320 |
0.2466699268 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 8 |
Hb_001258_060 |
0.2470814881 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 9 |
Hb_001565_110 |
0.2522533241 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 10 |
Hb_000414_110 |
0.2560412029 |
- |
- |
PREDICTED: MATE efflux family protein 5 isoform X1 [Populus euphratica] |
| 11 |
Hb_000318_200 |
0.2565328783 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 12 |
Hb_004242_190 |
0.261106304 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 13 |
Hb_003073_180 |
0.2642826261 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 14 |
Hb_000260_290 |
0.2666354249 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_080147_100 |
0.2674875986 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN [Jatropha curcas] |
| 16 |
Hb_138585_110 |
0.2749110358 |
- |
- |
- |
| 17 |
Hb_014894_010 |
0.2774573038 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 18 |
Hb_008864_090 |
0.2803831982 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 19 |
Hb_007763_030 |
0.2822201655 |
- |
- |
PREDICTED: uncharacterized protein LOC105638154 [Jatropha curcas] |
| 20 |
Hb_001507_100 |
0.2836319918 |
- |
- |
cytochrome P450, putative [Ricinus communis] |