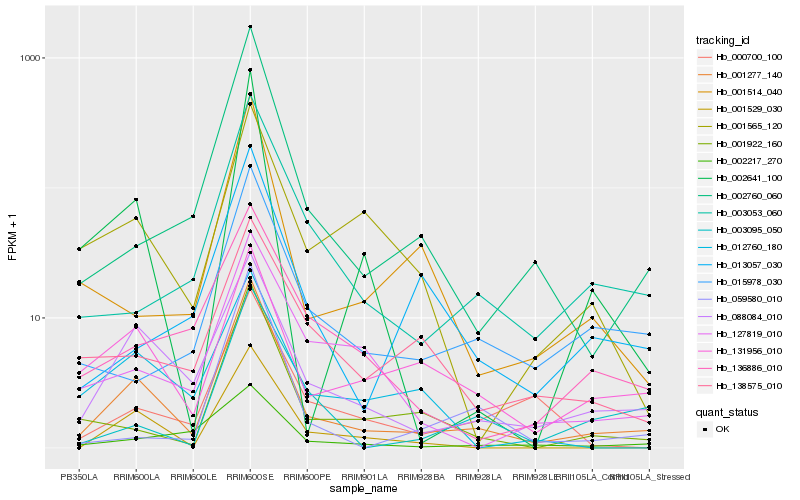
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_140 |
0.0 |
- |
- |
PREDICTED: plasma membrane calcium-transporting ATPase 2-like [Takifugu rubripes] |
| 2 |
Hb_088084_010 |
0.1521269609 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 3 |
Hb_001529_030 |
0.1891189456 |
- |
- |
hypothetical protein PRUPE_ppa024472mg, partial [Prunus persica] |
| 4 |
Hb_002760_060 |
0.1943215707 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 5 |
Hb_001922_160 |
0.2012874732 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 6 |
Hb_131956_010 |
0.2016582021 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 7 |
Hb_003053_060 |
0.2024297018 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 8 |
Hb_002641_100 |
0.2095105319 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 9 |
Hb_015978_030 |
0.218762196 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 10 |
Hb_001514_040 |
0.2216232928 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 11 |
Hb_002217_270 |
0.2247455188 |
- |
- |
PREDICTED: desiccation-related protein PCC13-62-like [Jatropha curcas] |
| 12 |
Hb_136886_010 |
0.2258452348 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 13 |
Hb_003095_050 |
0.2259512215 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 14 |
Hb_127819_010 |
0.2259798007 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 15 |
Hb_013057_030 |
0.2260835547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_059580_010 |
0.2270773425 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 17 |
Hb_000700_100 |
0.2292412681 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 18 |
Hb_001565_120 |
0.2377726002 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_138575_010 |
0.2394970296 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 20 |
Hb_012760_180 |
0.2399227064 |
- |
- |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |