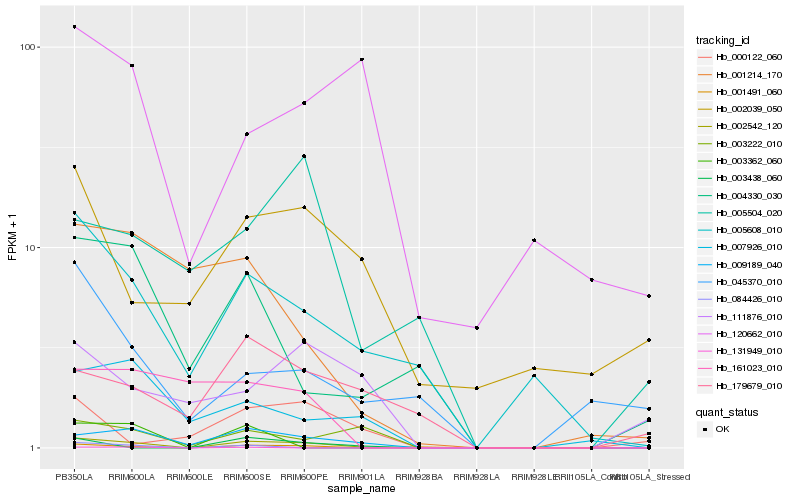
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002542_120 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 2 |
Hb_009189_040 |
0.2351453692 |
- |
- |
PREDICTED: uncharacterized protein LOC104446949 [Eucalyptus grandis] |
| 3 |
Hb_001491_060 |
0.2547032064 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6-like isoform X1 [Populus euphratica] |
| 4 |
Hb_005608_010 |
0.2715902763 |
- |
- |
PREDICTED: U-box domain-containing protein 40 [Jatropha curcas] |
| 5 |
Hb_003222_010 |
0.3028788876 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 6 |
Hb_007926_010 |
0.3139108922 |
- |
- |
PREDICTED: UPF0554 protein C2orf43 homolog isoform X4 [Citrus sinensis] |
| 7 |
Hb_004330_030 |
0.331266661 |
- |
- |
- |
| 8 |
Hb_003438_060 |
0.3372234144 |
- |
- |
- |
| 9 |
Hb_179679_010 |
0.3379939052 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_084426_010 |
0.3432229991 |
- |
- |
- |
| 11 |
Hb_001214_170 |
0.3488119843 |
- |
- |
pyruvate kinase family protein [Medicago truncatula] |
| 12 |
Hb_161023_010 |
0.3491840103 |
- |
- |
PREDICTED: uncharacterized protein LOC105639929 [Jatropha curcas] |
| 13 |
Hb_002039_050 |
0.3509876802 |
- |
- |
PREDICTED: U-box domain-containing protein 40 [Jatropha curcas] |
| 14 |
Hb_000122_060 |
0.351964201 |
- |
- |
PREDICTED: protein LURP-one-related 14 [Jatropha curcas] |
| 15 |
Hb_045370_010 |
0.3539915669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003362_060 |
0.3545611809 |
- |
- |
- |
| 17 |
Hb_120662_010 |
0.3554250046 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 18 |
Hb_111876_010 |
0.3566582901 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 19 |
Hb_131949_010 |
0.3588839185 |
- |
- |
PREDICTED: uncharacterized protein LOC104227863, partial [Nicotiana sylvestris] |
| 20 |
Hb_005504_020 |
0.3627796986 |
- |
- |
- |