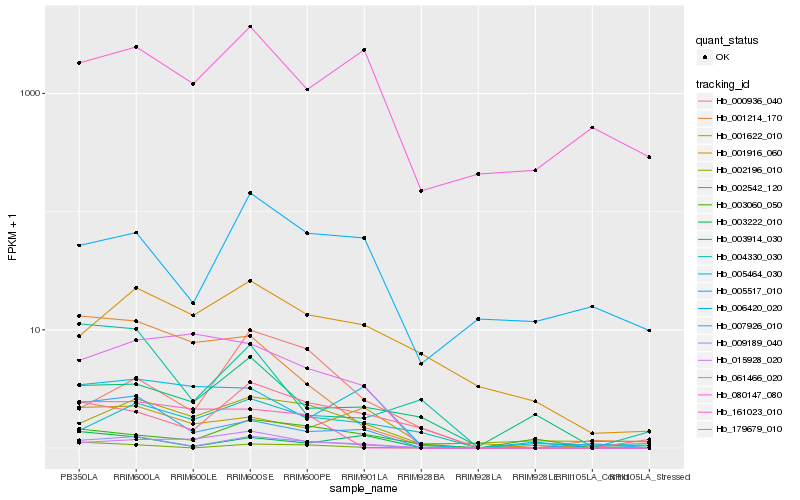
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009189_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104446949 [Eucalyptus grandis] |
| 2 |
Hb_007926_010 |
0.2243424167 |
- |
- |
PREDICTED: UPF0554 protein C2orf43 homolog isoform X4 [Citrus sinensis] |
| 3 |
Hb_002542_120 |
0.2351453692 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 4 |
Hb_015928_020 |
0.2402830758 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 5 |
Hb_179679_010 |
0.2587472631 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_003222_010 |
0.2603242533 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 7 |
Hb_002196_010 |
0.2725325347 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: proline-rich receptor-like protein kinase PERK1 [Jatropha curcas] |
| 8 |
Hb_061466_020 |
0.2751721033 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 9 |
Hb_000936_040 |
0.2794809495 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 10 |
Hb_001214_170 |
0.2868989587 |
- |
- |
pyruvate kinase family protein [Medicago truncatula] |
| 11 |
Hb_161023_010 |
0.2905488296 |
- |
- |
PREDICTED: uncharacterized protein LOC105639929 [Jatropha curcas] |
| 12 |
Hb_006420_020 |
0.2910687502 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 13 |
Hb_003060_050 |
0.2943412562 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 14 |
Hb_001622_010 |
0.296075547 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_005464_030 |
0.2966995768 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_005517_010 |
0.2981713909 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001916_060 |
0.2991732342 |
- |
- |
PREDICTED: putative protein FAR1-RELATED SEQUENCE 10 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003914_030 |
0.299324478 |
- |
- |
- |
| 19 |
Hb_004330_030 |
0.3035984939 |
- |
- |
- |
| 20 |
Hb_080147_080 |
0.3040361645 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |