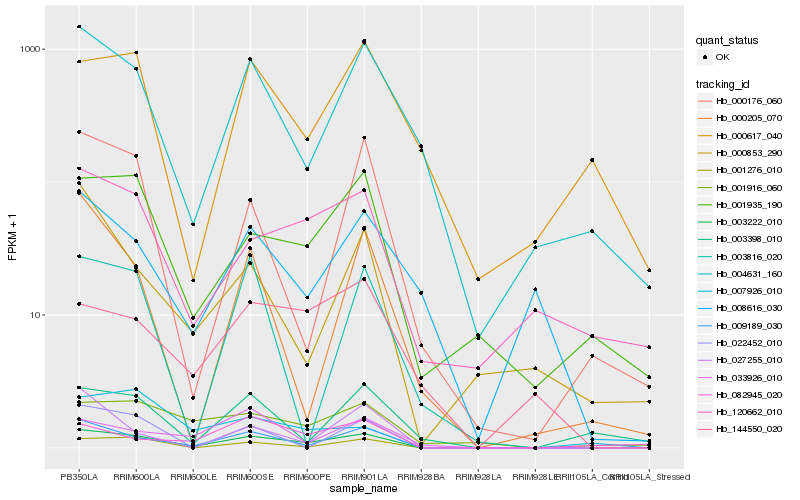
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003222_010 |
0.0 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 2 |
Hb_009189_030 |
0.1427549299 |
- |
- |
hypothetical protein PRUPE_ppa023969mg, partial [Prunus persica] |
| 3 |
Hb_001276_010 |
0.1833260289 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_001935_190 |
0.1986481449 |
- |
- |
Nuc-1 negative regulatory protein preg, putative [Ricinus communis] |
| 5 |
Hb_004631_160 |
0.2084028821 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 6 |
Hb_120662_010 |
0.221900403 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 7 |
Hb_001916_060 |
0.2276543382 |
- |
- |
PREDICTED: putative protein FAR1-RELATED SEQUENCE 10 isoform X2 [Jatropha curcas] |
| 8 |
Hb_144550_020 |
0.236331638 |
- |
- |
- |
| 9 |
Hb_000176_060 |
0.236515635 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_033926_010 |
0.2374498639 |
- |
- |
- |
| 11 |
Hb_022452_010 |
0.2384899544 |
- |
- |
- |
| 12 |
Hb_007926_010 |
0.2436470829 |
- |
- |
PREDICTED: UPF0554 protein C2orf43 homolog isoform X4 [Citrus sinensis] |
| 13 |
Hb_008616_030 |
0.2465608744 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003816_020 |
0.2495368578 |
- |
- |
hypothetical protein POPTR_0017s05940g [Populus trichocarpa] |
| 15 |
Hb_003398_010 |
0.2531242195 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Eucalyptus grandis] |
| 16 |
Hb_000617_040 |
0.2542784723 |
- |
- |
PREDICTED: uncharacterized protein LOC105647487 isoform X2 [Jatropha curcas] |
| 17 |
Hb_027255_010 |
0.2548646578 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000853_290 |
0.2556329471 |
- |
- |
PREDICTED: uncharacterized protein LOC105642585 [Jatropha curcas] |
| 19 |
Hb_082945_020 |
0.2559038519 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 20 |
Hb_000205_070 |
0.2560594241 |
- |
- |
kinase, putative [Ricinus communis] |