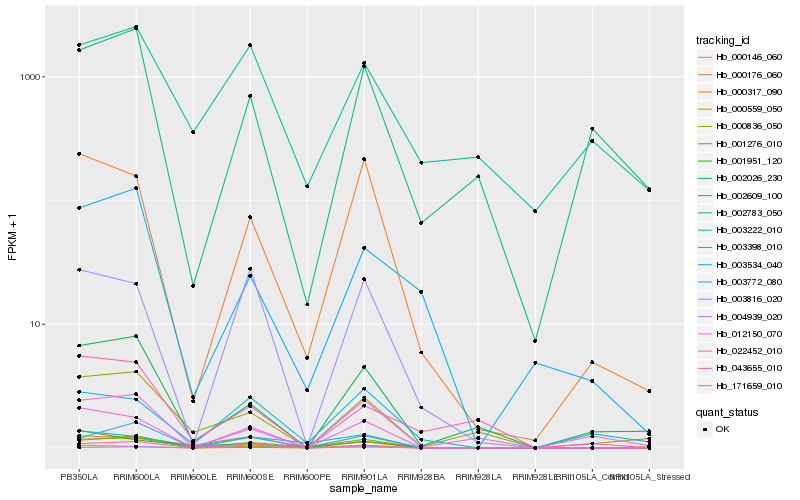
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171659_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104903214 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_003772_080 |
0.1513675868 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 3 |
Hb_022452_010 |
0.1588110981 |
- |
- |
- |
| 4 |
Hb_001276_010 |
0.1664682774 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000317_090 |
0.1778566327 |
- |
- |
hypothetical protein RCOM_1176360 [Ricinus communis] |
| 6 |
Hb_003816_020 |
0.1782075197 |
- |
- |
hypothetical protein POPTR_0017s05940g [Populus trichocarpa] |
| 7 |
Hb_004939_020 |
0.2038312513 |
- |
- |
gag/pol protein [Bryonia dioica] |
| 8 |
Hb_000176_060 |
0.255984034 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000146_060 |
0.2626238647 |
- |
- |
- |
| 10 |
Hb_002026_230 |
0.2647270854 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_000836_050 |
0.2675707355 |
- |
- |
- |
| 12 |
Hb_002609_100 |
0.2696803053 |
- |
- |
PREDICTED: uncharacterized protein LOC105133073 isoform X1 [Populus euphratica] |
| 13 |
Hb_003534_040 |
0.2709358501 |
- |
- |
hypothetical protein POPTR_0009s14420g [Populus trichocarpa] |
| 14 |
Hb_000559_050 |
0.2807522053 |
- |
- |
hypothetical protein JCGZ_20429 [Jatropha curcas] |
| 15 |
Hb_003222_010 |
0.2833235105 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 16 |
Hb_003398_010 |
0.2851219264 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Eucalyptus grandis] |
| 17 |
Hb_001951_120 |
0.2896665922 |
- |
- |
- |
| 18 |
Hb_012150_070 |
0.2917350686 |
- |
- |
hypothetical protein JCGZ_04984 [Jatropha curcas] |
| 19 |
Hb_043655_010 |
0.291941453 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 20 |
Hb_002783_050 |
0.2930095414 |
- |
- |
zinc finger protein, putative [Ricinus communis] |