| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
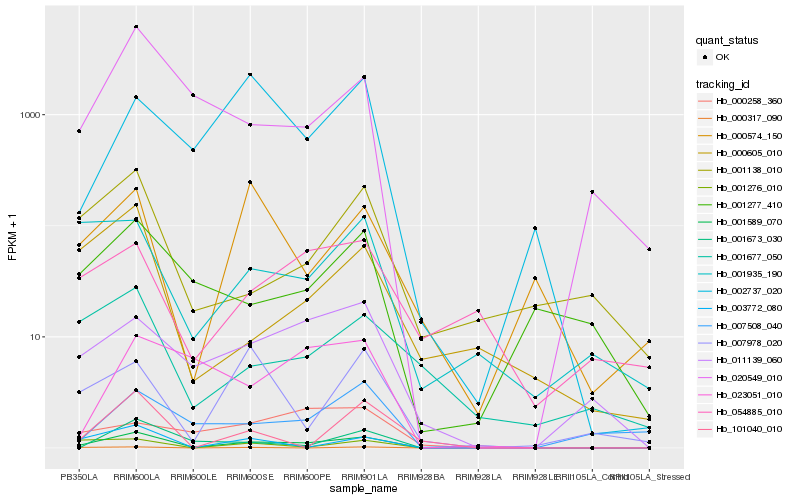
Hb_001589_070 |
0.0 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 2 |
Hb_011139_060 |
0.2521374985 |
- |
- |
- |
| 3 |
Hb_003772_080 |
0.252788975 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 4 |
Hb_020549_010 |
0.2630598341 |
- |
- |
PREDICTED: uncharacterized protein LOC105638664 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001673_030 |
0.2864873499 |
- |
- |
hypothetical protein JCGZ_08729 [Jatropha curcas] |
| 6 |
Hb_002737_020 |
0.2892098174 |
- |
- |
metallothionein [Bruguiera gymnorhiza] |
| 7 |
Hb_001935_190 |
0.2915975104 |
- |
- |
Nuc-1 negative regulatory protein preg, putative [Ricinus communis] |
| 8 |
Hb_001276_010 |
0.2940389227 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_001277_410 |
0.3001253908 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 10 |
Hb_001138_010 |
0.3020246301 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000317_090 |
0.3026643542 |
- |
- |
hypothetical protein RCOM_1176360 [Ricinus communis] |
| 12 |
Hb_007508_040 |
0.3035480895 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001677_050 |
0.3041979636 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP8, putative [Ricinus communis] |
| 14 |
Hb_101040_010 |
0.3053048895 |
- |
- |
- |
| 15 |
Hb_000605_010 |
0.3054928596 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 16 |
Hb_023051_010 |
0.3061073705 |
- |
- |
PREDICTED: putative amidase C869.01 [Populus euphratica] |
| 17 |
Hb_000574_150 |
0.3062244303 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 18 |
Hb_000258_360 |
0.3072919956 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 19 |
Hb_007978_020 |
0.308628837 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA1-like [Jatropha curcas] |
| 20 |
Hb_054885_010 |
0.3102234067 |
- |
- |
- |